Related Research Articles

In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed from an RNA. cDNA exists in both single-stranded and double-stranded forms and in both natural and engineered forms.

In biology, translation is the process in living cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotides in the RNA. The nucleotides are considered three at a time. Each such triple results in addition of one specific amino acid to the protein being generated. The matching from nucleotide triple to amino acid is called the genetic code. The translation is performed by a large complex of functional RNA and proteins called ribosomes. The entire process is called gene expression.
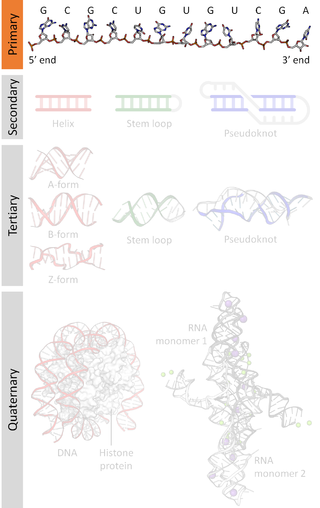
A nucleic acid sequence is a succession of bases within the nucleotides forming alleles within a DNA or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the order of the nucleotides. By convention, sequences are usually presented from the 5' end to the 3' end. For DNA, with its double helix, there are two possible directions for the notated sequence; of these two, the sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the primary structure.

Transfer RNA is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length, that serves as the physical link between the mRNA and the amino acid sequence of proteins. Transfer RNA (tRNA) does this by carrying an amino acid to the protein-synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code.

Functional genomics is a field of molecular biology that attempts to describe gene functions and interactions. Functional genomics make use of the vast data generated by genomic and transcriptomic projects. Functional genomics focuses on the dynamic aspects such as gene transcription, translation, regulation of gene expression and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional "candidate-gene" approach.
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The term transcriptome is a portmanteau of the words transcript and genome; it is associated with the process of transcript production during the biological process of transcription.

RNA editing is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms and is one of the most evolutionarily conserved properties of RNAs. RNA editing may include the insertion, deletion, and base substitution of nucleotides within the RNA molecule. RNA editing is relatively rare, with common forms of RNA processing not usually considered as editing. It can affect the activity, localization as well as stability of RNAs, and has been linked with human diseases.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recapping.
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global binding sites precisely for any protein of interest. Previously, ChIP-on-chip was the most common technique utilized to study these protein–DNA relations.

RNA-Seq is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome.

Ribosomal pause refers to the queueing or stacking of ribosomes during translation of the nucleotide sequence of mRNA transcripts. These transcripts are decoded and converted into an amino acid sequence during protein synthesis by ribosomes. Due to the pause sites of some mRNA's, there is a disturbance caused in translation. Ribosomal pausing occurs in both eukaryotes and prokaryotes. A more severe pause is known as a ribosomal stall.
Chimeric RNA, sometimes referred to as a fusion transcript, is composed of exons from two or more different genes that have the potential to encode novel proteins. These mRNAs are different from those produced by conventional splicing as they are produced by two or more gene loci.
Translation complex profile sequencing (TCP-seq) is a molecular biology method for obtaining snapshots of momentary distribution of protein synthesis complexes along messenger RNA (mRNA) chains.

In epitranscriptomic sequencing, most methods focus on either (1) enrichment and purification of the modified RNA molecules before running on the RNA sequencer, or (2) improving or modifying bioinformatics analysis pipelines to call the modification peaks. Most methods have been adapted and optimized for mRNA molecules, except for modified bisulfite sequencing for profiling 5-methylcytidine which was optimized for tRNAs and rRNAs.
Transcriptomics technologies are the techniques used to study an organism's transcriptome, the sum of all of its RNA transcripts. The information content of an organism is recorded in the DNA of its genome and expressed through transcription. Here, mRNA serves as a transient intermediary molecule in the information network, whilst non-coding RNAs perform additional diverse functions. A transcriptome captures a snapshot in time of the total transcripts present in a cell. Transcriptomics technologies provide a broad account of which cellular processes are active and which are dormant. A major challenge in molecular biology is to understand how a single genome gives rise to a variety of cells. Another is how gene expression is regulated.
Time-resolved RNA sequencing methods are applications of RNA-seq that allow for observations of RNA abundances over time in a biological sample or samples. Second-Generation DNA sequencing has enabled cost effective, high throughput and unbiased analysis of the transcriptome. Normally, RNA-seq is only capable of capturing a snapshot of the transcriptome at the time of sample collection. This necessitates multiple samplings at multiple time points, which increases both monetary and time costs for experiments. Methodological and technological innovations have allowed for the analysis of the RNA transcriptome over time without requiring multiple samplings at various time points.

Micropeptides are polypeptides with a length of less than 100-150 amino acids that are encoded by short open reading frames (sORFs). In this respect, they differ from many other active small polypeptides, which are produced through the posttranslational cleavage of larger polypeptides. In terms of size, micropeptides are considerably shorter than "canonical" proteins, which have an average length of 330 and 449 amino acids in prokaryotes and eukaryotes, respectively. Micropeptides are sometimes named according to their genomic location. For example, the translated product of an upstream open reading frame (uORF) might be called a uORF-encoded peptide (uPEP). Micropeptides lack an N-terminal signaling sequences, suggesting that they are likely to be localized to the cytoplasm. However, some micropeptides have been found in other cell compartments, as indicated by the existence of transmembrane micropeptides. They are found in both prokaryotes and eukaryotes. The sORFs from which micropeptides are translated can be encoded in 5' UTRs, small genes, or polycistronic mRNAs. Some micropeptide-coding genes were originally mis-annotated as long non-coding RNAs (lncRNAs).
Small proteins are a diverse fold class of proteins. Their tertiary structure is usually maintained by disulphide bridges, metal ligands, and or cofactors such as heme. Some small proteins serve important regulatory functions by direct interaction with certain enzymes and are therefore also an interesting tool for biotechnological applications in microorganisms.

Translatomics is the study of all open reading frames (ORFs) that are being actively translated in a cell or organism. This collection of ORFs is called the translatome. Characterizing a cell's translatome can give insight into the array of biological pathways that are active in the cell. According to the central dogma of molecular biology, the DNA in a cell is transcribed to produce RNA, which is then translated to produce a protein. Thousands of proteins are encoded in an organism's genome, and the proteins present in a cell cooperatively carry out many functions to support the life of the cell. Under various conditions, such as during stress or specific timepoints in development, the cell may require different biological pathways to be active, and therefore require a different collection of proteins. Depending on intrinsic and environmental conditions, the collection of proteins being made at one time varies. Translatomic techniques can be used to take a "snapshot" of this collection of actively translating ORFs, which can give information about which biological pathways the cell is activating under the present conditions.
References
- 1 2 3 4 5 6 7 8 Ingolia NT (March 2014). "Ribosome profiling: new views of translation, from single codons to genome scale". Nature Reviews. Genetics. 15 (3): 205–13. doi:10.1038/nrg3645. PMID 24468696. S2CID 13069682.
- 1 2 Dougherty, Joseph D. (13 December 2017). "The Expanding Toolkit of Translating Ribosome Affinity Purification". Journal of Neuroscience. 37 (50): 12079–12087. doi: 10.1523/JNEUROSCI.1929-17.2017 . PMC 5729187 . PMID 29237735.
- ↑ Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE; et al. (2008). "A translational profiling approach for the molecular characterization of CNS cell types". Cell. 135 (4): 738–48. doi:10.1016/j.cell.2008.10.028. PMC 2696821 . PMID 19013281.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - 1 2 Weiss RB, Atkins JF (December 2011). "Molecular biology. Translation goes global". Science. 334 (6062): 1509–10. doi:10.1126/science.1216974. PMID 22174241. S2CID 206538968.
- 1 2 3 Michel AM, Baranov PV (September 2013). "Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale". Wiley Interdisciplinary Reviews: RNA. 4 (5): 473–90. doi:10.1002/wrna.1172. PMC 3823065 . PMID 23696005.
- 1 2 Buskirk AR, Green R (March 2017). "Ribosome pausing, arrest and rescue in bacteria and eukaryotes". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 372 (1716): 20160183. doi:10.1098/rstb.2016.0183. PMC 5311927 . PMID 28138069.
- 1 2 3 Andreev DE, O'Connor PB, Loughran G, Dmitriev SE, Baranov PV, Shatsky IN (January 2017). "Insights into the mechanisms of eukaryotic translation gained with ribosome profiling". Nucleic Acids Research. 45 (2): 513–526. doi:10.1093/nar/gkw1190. PMC 5314775 . PMID 27923997.
- ↑ Ozadam, Hakan; Geng, Michael; Cenik, Can (1 May 2020). "RiboFlow, RiboR and RiboPy: an ecosystem for analyzing ribosome profiling data at read length resolution". Bioinformatics. 36 (9): 2929–2931. doi:10.1093/bioinformatics/btaa028. ISSN 1367-4811. PMC 7203755 . PMID 31930375.
- ↑ Oertlin, Christian; Lorent, Julie; Murie, Carl; Furic, Luc; Topisirovic, Ivan; Larsson, Ola (9 July 2019). "Generally applicable transcriptome-wide analysis of translation using anota2seq". Nucleic Acids Research. 47 (12): e70. doi:10.1093/nar/gkz223. ISSN 1362-4962. PMC 6614820 . PMID 30926999.
- ↑ Cenik, Can; Cenik, Elif Sarinay; Byeon, Gun W.; Grubert, Fabian; Candille, Sophie I.; Spacek, Damek; Alsallakh, Bilal; Tilgner, Hagen; Araya, Carlos L.; Tang, Hua; Ricci, Emiliano (November 2015). "Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans". Genome Research. 25 (11): 1610–1621. doi:10.1101/gr.193342.115. ISSN 1549-5469. PMC 4617958 . PMID 26297486.
- 1 2 Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (April 2009). "Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling". Science. 324 (5924): 218–23. Bibcode:2009Sci...324..218I. doi:10.1126/science.1168978. PMC 2746483 . PMID 19213877.
- ↑ Ozadam, Hakan; Geng, Michael; Cenik, Can (1 May 2020). "RiboFlow, RiboR and RiboPy: an ecosystem for analyzing ribosome profiling data at read length resolution". Bioinformatics. 36 (9): 2929–2931. doi:10.1093/bioinformatics/btaa028. ISSN 1367-4811. PMC 7203755 . PMID 31930375.