Related Research Articles

Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis is induced. The system involves the RecA protein. The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (LexA) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages. This can eventually lead to malignant tumors, or cancer as per the two hit hypothesis.
Integrons are genetic mechanisms that allow bacteria to adapt and evolve rapidly through the stockpiling and expression of new genes. These genes are embedded in a specific genetic structure called gene cassette that generally carries one promoterless open reading frame (ORF) together with a recombination site (attC). Integron cassettes are incorporated to the attI site of the integron platform by site-specific recombination reactions mediated by the integrase.
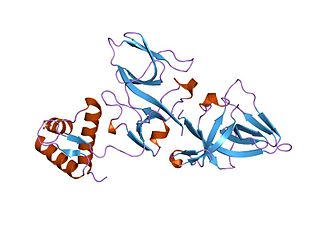
Repressor LexA or LexA is a transcriptional repressor that represses SOS response genes coding primarily for error-prone DNA polymerases, DNA repair enzymes and cell division inhibitors. LexA forms de facto a two-component regulatory system with RecA, which senses DNA damage at stalled replication forks, forming monofilaments and acquiring an active conformation capable of binding to LexA and causing LexA to cleave itself, in a process called autoproteolysis.

The TATA-binding protein (TBP) is a general transcription factor that binds specifically to a DNA sequence called the TATA box. This DNA sequence is found about 30 base pairs upstream of the transcription start site in some eukaryotic gene promoters.
The adaptive response is a form of direct DNA repair in E. coli that protects DNA from damage by external agents or by errors during replication. It is initiated against alkylation, particularly methylation, of guanine or thymine nucleotides or phosphate groups on the sugar-phosphate backbone of DNA. Under sustained exposure to low-level treatment with alkylating mutagens, E. coli can adapt to the presence of the mutagen, rendering subsequent treatment with high doses of the same agent less effective.
In molecular genetics, a regulon is a group of genes that are regulated as a unit, generally controlled by the same regulatory gene that expresses a protein acting as a repressor or activator. This terminology is generally, although not exclusively, used in reference to prokaryotes, whose genomes are often organized into operons; the genes contained within a regulon are usually organized into more than one operon at disparate locations on the chromosome. Applied to eukaryotes, the term refers to any group of non-contiguous genes controlled by the same regulatory gene.
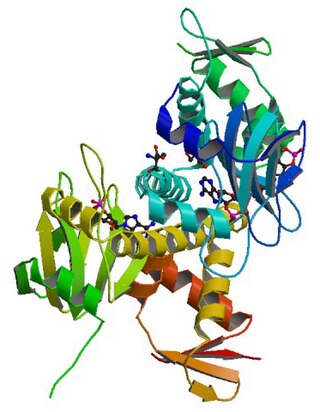
cAMP receptor protein is a regulatory protein in bacteria. CRP protein binds cAMP, which causes a conformational change that allows CRP to bind tightly to a specific DNA site in the promoters of the genes it controls. CRP then activates transcription through direct protein–protein interactions with RNA polymerase.

Y box binding protein 1 also known as Y-box transcription factor or nuclease-sensitive element-binding protein 1 is a protein that in humans is encoded by the YBX1 gene.
In biology, phase variation is a method for dealing with rapidly varying environments without requiring random mutation. It involves the variation of protein expression, frequently in an on-off fashion, within different parts of a bacterial population. As such the phenotype can switch at frequencies that are much higher than classical mutation rates. Phase variation contributes to virulence by generating heterogeneity. Although it has been most commonly studied in the context of immune evasion, it is observed in many other areas as well and is employed by various types of bacteria, including Salmonella species.

The JUMPstart RNA motif describes a conserved RNA-based secondary structure associated with JUMPstart elements. The 39-base-pair JUMPstart sequence describes a conserved element upstream of genes that participate in polysaccharide synthesis. The JUMPstart element has been shown to function as an RNA, and is present in the 5' untranslated regions of the genes it regulates.

The rsmX gene is part of the Rsm/Csr family of non-coding RNAs (ncRNAs). Members of the Rsm/Csr family are present in a diverse range of bacteria, including Escherichia coli, Erwinia, Salmonella, Vibrio and Pseudomonas. These ncRNAs act by sequestering translational repressor proteins, called RsmA, activating expression of downstream genes that would normally be blocked by the repressors. Sequestering of target proteins is dependent upon exposed GGA motifs in the stem loops of the ncRNAs. Typically, the activated genes are involved in secondary metabolism, biofilm formation and motility.

FnrS RNA is a family of Hfq-binding small RNA whose expression is upregulated in response to anaerobic conditions. It is named FnrS because its expression is strongly dependent on fumarate and nitrate reductase regulator (FNR), a direct oxygen availability sensor.

The TisB-IstR toxin-antitoxin system is the first known toxin-antitoxin system which is induced by the SOS response in response to DNA damage.
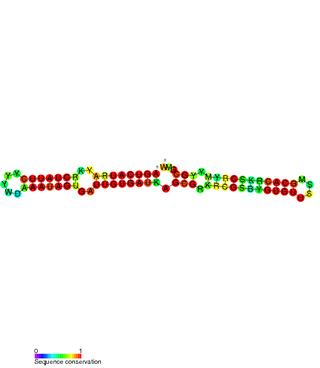
The SymE-SymR toxin-antitoxin system consists of a small symbiotic endonuclease toxin, SymE, and a non-coding RNA symbiotic RNA antitoxin, SymR, which inhibits SymE translation. SymE-SymR is a type I toxin-antitoxin system, and is under regulation by the antitoxin, SymR. The SymE-SymR complex is believed to play an important role in recycling damaged RNA and DNA. The relationship and corresponding structures of SymE and SymR provide insight into the mechanism of toxicity and overall role in prokaryotic systems.

In molecular biology, the LuxR-type DNA-binding HTH domain is a DNA-binding, helix-turn-helix (HTH) domain of about 65 amino acids. It is present in transcription regulators of the LuxR/FixJ family of response regulators. The domain is named after Vibrio fischeri luxR, a transcriptional activator for quorum-sensing control of luminescence. LuxR-type HTH domain proteins occur in a variety of organisms. The DNA-binding HTH domain is usually located in the C-terminal region of the protein; the N-terminal region often containing an autoinducer-binding domain or a response regulatory domain. Most luxR-type regulators act as transcription activators, but some can be repressors or have a dual role for different sites. LuxR-type HTH regulators control a wide variety of activities in various biological processes.
Oxidation response is stimulated by a disturbance in the balance between the production of reactive oxygen species and antioxidant responses, known as oxidative stress. Active species of oxygen naturally occur in aerobic cells and have both intracellular and extracellular sources. These species, if not controlled, damage all components of the cell, including proteins, lipids and DNA. Hence cells need to maintain a strong defense against the damage. The following table gives an idea of the antioxidant defense system in bacterial system.
The gene rpoN encodes the sigma factor sigma-54, a protein in Escherichia coli and other species of bacteria. RpoN antagonizes RpoS sigma factors.

Ivan Erill is a Spanish computational biologist known for his research in comparative genomics and molecular microbiology. His work focuses primarily on bacterial comparative genomics, through the development of computational methods for analyzing regulatory networks and their evolution.
References
- ↑ Walker, GC (October 1995). "SOS-regulated proteins in translesion DNA synthesis and mutagenesis". Trends in Biochemical Sciences. 20 (10): 416–20. doi:10.1016/s0968-0004(00)89091-x. PMID 8533155.
- ↑ Gillor, Osnat; Vriezen, Jan A. C.; Riley, Margaret A. (Jun 2008). "The role of SOS boxes in enteric bacteriocin regulation". Microbiology. 154 (6): 1783–1792. doi:10.1099/mic.0.2007/016139-0. PMC 2729051 . PMID 18524933.
- ↑ Fernández de Henestrosa AR, Rivera E, Tapias A, Barbé J (June 1998). "Identification of the Rhodobacter sphaeroides SOS box". Mol. Microbiol. 28 (5): 991–1003. doi: 10.1046/j.1365-2958.1998.00860.x . PMID 9663685.
- ↑ Tapias A, Barbé J (August 1999). "Regulation of divergent transcription from the uvrA-ssb promoters in Sinorhizobium meliloti". Mol. Gen. Genet. 262 (1): 121–30. doi:10.1007/s004380051066. PMID 10503543. S2CID 2373618.
- ↑ Erill I, Escribano M, Campoy S, Barbé J (November 2003). "In silico analysis reveals substantial variability in the gene contents of the gamma proteobacteria LexA-regulon". Bioinformatics. 19 (17): 2225–36. doi: 10.1093/bioinformatics/btg303 . PMID 14630651.
- ↑ Campoy S, Fontes M, Padmanabhan S, Cortés P, Llagostera M, Barbé J (August 2003). "LexA-independent DNA damage-mediated induction of gene expression in Myxococcus xanthus". Mol. Microbiol. 49 (3): 769–81. doi:10.1046/j.1365-2958.2003.03592.x. PMID 12864858.
- ↑ Winterling KW, Chafin D, Hayes JJ, et al. (15 April 1998). "The Bacillus subtilis DinR Binding Site: Redefinition of the Consensus Sequence". J. Bacteriol. 180 (8): 2201–11. doi:10.1128/JB.180.8.2201-2211.1998. PMC 107149 . PMID 9555905.
- ↑ Davis EO, Dullaghan EM, Rand L (June 2002). "Definition of the Mycobacterial SOS Box and Use To Identify LexA-Regulated Genes in Mycobacterium tuberculosis". J. Bacteriol. 184 (12): 3287–95. doi:10.1128/JB.184.12.3287-3295.2002. PMC 135081 . PMID 12029045.
- ↑ Mazón G, Lucena JM, Campoy S, Fernández de Henestrosa AR, Candau P, Barbé J (February 2004). "LexA-binding sequences in Gram-positive and cyanobacteria are closely related". Mol. Genet. Genomics. 271 (1): 40–9. doi:10.1007/s00438-003-0952-x. PMID 14652736. S2CID 9219764.
- Erill I, et al. (2004). "Differences in LexA regulon structure among Proteobacteria through in vivo assisted comparative genomics". Nucleic Acids Research. 32 (22): 6617–26. doi:10.1093/nar/gkh996. PMC 545464 . PMID 15604457.
- Shinagawa H (1996). "SOS response as an adaptive response to DNA damage in prokaryotes". Stress-Inducible Cellular Responses. pp. 221–35. doi:10.1007/978-3-0348-9088-5_14. ISBN 978-3-0348-9901-7. PMID 8856977.
{{cite book}}:|journal=ignored (help)