
Escherichia coli ( ESH-ə-RIK-ee-ə KOH-ly) is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes such as EPEC, and ETEC are pathogenic and can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains are part of the normal microbiota of the gut and are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of E. coli benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between E. coli and humans are a type of mutualistic biological relationship — where both the humans and the E. coli are benefitting each other. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.

Salmonella is a genus of rod-shaped (bacillus) gram-negative bacteria of the family Enterobacteriaceae. The two known species of Salmonella are Salmonella enterica and Salmonella bongori. S. enterica is the type species and is further divided into six subspecies that include over 2,600 serotypes. Salmonella was named after Daniel Elmer Salmon (1850–1914), an American veterinary surgeon.

Shigella is a genus of bacteria that is Gram-negative, facultatively anaerobic, non–spore-forming, nonmotile, rod-shaped, and is genetically closely related to Escherichia. The genus is named after Kiyoshi Shiga, who discovered it in 1897.

Shiga toxins are a family of related toxins with two major groups, Stx1 and Stx2, expressed by genes considered to be part of the genome of lambdoid prophages. The toxins are named after Kiyoshi Shiga, who first described the bacterial origin of dysentery caused by Shigella dysenteriae. Shiga-like toxin (SLT) is a historical term for similar or identical toxins produced by Escherichia coli. The most common sources for Shiga toxin are the bacteria S. dysenteriae and some serotypes of Escherichia coli (STEC), which includes serotypes O157:H7, and O104:H4.

Salmonella enterica is a rod-shaped, flagellate, facultative anaerobic, Gram-negative bacterium and a species of the genus Salmonella. It is divided into six subspecies, arizonae (IIIa), diarizonae (IIIb), houtenae (IV), salamae (II), indica (VI), and enterica (I). A number of its serovars are serious human pathogens; many of them are serovars of Salmonella enterica subsp. enterica.

The bacterial capsule is a large structure common to many bacteria. It is a polysaccharide layer that lies outside the cell envelope, and is thus deemed part of the outer envelope of a bacterial cell. It is a well-organized layer, not easily washed off, and it can be the cause of various diseases.
Bacillary dysentery is a type of dysentery, and is a severe form of shigellosis. It is associated with species of bacteria from the family Enterobacteriaceae. The term is usually restricted to Shigella infections.

Shigella flexneri is a species of Gram-negative bacteria in the genus Shigella that can cause diarrhea in humans. Several different serogroups of Shigella are described; S. flexneri belongs to group B. S. flexneri infections can usually be treated with antibiotics, although some strains have become resistant. Less severe cases are not usually treated because they become more resistant in the future. Shigella are closely related to Escherichia coli, but can be differentiated from E.coli based on pathogenicity, physiology and serology.
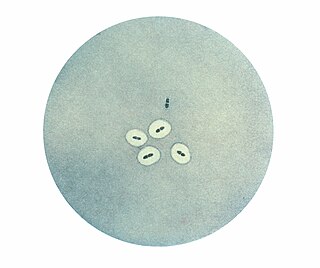
The quellung reaction, also called the Neufeld reaction, is a biochemical reaction in which antibodies bind to the bacterial capsule of Streptococcus pneumoniae, Klebsiella pneumoniae, Neisseria meningitidis, Bacillus anthracis, Haemophilus influenzae, Escherichia coli, and Salmonella. The antibody reaction allows these species to be visualized under a microscope. If the reaction is positive, the capsule becomes opaque and appears to enlarge.
The AB5 toxins are six-component protein complexes secreted by certain pathogenic bacteria known to cause human diseases such as cholera, dysentery, and hemolytic–uremic syndrome. One component is known as the A subunit, and the remaining five components are B subunits. All of these toxins share a similar structure and mechanism for entering targeted host cells. The B subunit is responsible for binding to receptors to open up a pathway for the A subunit to enter the cell. The A subunit is then able to use its catalytic machinery to take over the host cell's regular functions.

Leucine responsive protein, or Lrp, is a global regulator protein, meaning that it regulates the biosynthesis of leucine, as well as the other branched-chain amino acids, valine and isoleucine. In bacteria, it is encoded by the lrp gene.

Hektoen enteric agar is a selective and differential agar primarily used to recover Salmonella and Shigella from patient specimens. HEA contains indicators of lactose fermentation and hydrogen sulfide production; as well as inhibitors to prevent the growth of Gram-positive bacteria. It is named after the Hektoen Institute in Chicago, where researchers developed the agar.

Viable but nonculturable (VBNC) bacteria refers as to bacteria that are in a state of very low metabolic activity and do not divide, but are alive and have the ability to become culturable once resuscitated.

Salmonella enterica subsp. enterica is a subspecies of Salmonella enterica, the rod-shaped, flagellated, aerobic, Gram-negative bacterium. Many of the pathogenic serovars of the S. enterica species are in this subspecies, including that responsible for typhoid.
Omptins are a family of bacterial proteases. They are aspartate proteases, which cleave peptides with the use of a water molecule. Found in the outer membrane of gram-negative enterobacteria such as Shigella flexneri, Yersinia pestis, Escherichia coli, and Salmonella enterica. Omptins consist of a widely conserved beta barrel spanning the membrane with 5 extracellular loops. These loops are responsible for the various substrate specificities. These proteases rely upon binding of lipopolysaccharide for activity.
Cytolethal distending toxins are a class of heterotrimeric toxins produced by certain gram-negative bacteria that display DNase activity. These toxins trigger G2/M cell cycle arrest in specific mammalian cell lines, leading to the enlarged or distended cells for which these toxins are named. Affected cells die by apoptosis.

Escherichia coli is a gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms (endotherms). Most E. coli strains are harmless, but pathogenic varieties cause serious food poisoning, septic shock, meningitis, or urinary tract infections in humans. Unlike normal flora E. coli, the pathogenic varieties produce toxins and other virulence factors that enable them to reside in parts of the body normally not inhabited by E. coli, and to damage host cells. These pathogenic traits are encoded by virulence genes carried only by the pathogens.
Bacterial effectors are proteins secreted by pathogenic bacteria into the cells of their host, usually using a type 3 secretion system (TTSS/T3SS), a type 4 secretion system (TFSS/T4SS) or a Type VI secretion system (T6SS). Some bacteria inject only a few effectors into their host’s cells while others may inject dozens or even hundreds. Effector proteins may have many different activities, but usually help the pathogen to invade host tissue, suppress its immune system, or otherwise help the pathogen to survive. Effector proteins are usually critical for virulence. For instance, in the causative agent of plague, the loss of the T3SS is sufficient to render the bacteria completely avirulent, even when they are directly introduced into the bloodstream. Gram negative microbes are also suspected to deploy bacterial outer membrane vesicles to translocate effector proteins and virulence factors via a membrane vesicle trafficking secretory pathway, in order to modify their environment or attack/invade target cells, for example, at the host-pathogen interface.

Edward Thomas Ryan is an American microbiologist, immunologist, and physician at Harvard University and Massachusetts General Hospital. Ryan served as president of the American Society of Tropical Medicine and Hygiene from 2009 to 2010. Ryan is Professor of Immunology and Infectious Diseases at the Harvard T.H. Chan School of Public Health, Professor of Medicine at Harvard Medical School, and Director of Global Infectious Diseases at the Massachusetts General Hospital. Ryan's research and clinical focus has been on infectious diseases associated with residing in, immigrating from, or traveling through resource-limited areas. Ryan is a Fellow of the American Society of Microbiology, the American Society of Tropical Medicine and Hygiene, the American College of Physicians, and the Infectious Diseases Society of America.

The mobilized colistin resistance (mcr) gene confers plasmid-mediated resistance to colistin, one of a number of last-resort antibiotics for treating Gram-negative infections. mcr-1, the original variant, is capable of horizontal transfer between different strains of a bacterial species. After discovery in November 2015 in E. coli from a pig in China it has been found in Escherichia coli, Salmonella enterica, Klebsiella pneumoniae, Enterobacter aerogenes, and Enterobacter cloacae. As of 2017, it has been detected in more than 30 countries on 5 continents in less than a year.















