Antioxidants are compounds that inhibit oxidation, a chemical reaction that can produce free radicals. Autoxidation leads to degradation of organic compounds, including living matter. Antioxidants are frequently added to industrial products, such as polymers, fuels, and lubricants, to extend their useable lifetimes. Food are also treated with antioxidants to forestall spoilage, in particular the rancidification of oils and fats. In cells, antioxidants such as glutathione, mycothiol or bacillithiol, and enzyme systems like superoxide dismutase, can prevent damage from oxidative stress.

Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.

An electron transport chain (ETC) is a series of protein complexes and other molecules that transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. The electrons that transferred from NADH and FADH2 to the ETC involves four multi-subunit large enzymes complexes and two mobile electron carriers. Many of the enzymes in the electron transport chain are membrane-bound.
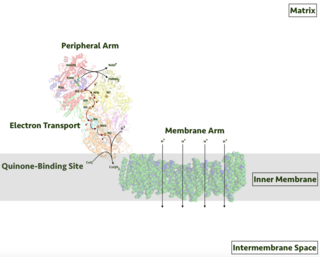
Respiratory complex I, EC 7.1.1.2 is the first large protein complex of the respiratory chains of many organisms from bacteria to humans. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and translocates protons across the inner mitochondrial membrane in eukaryotes or the plasma membrane of bacteria.

The coenzyme Q : cytochrome c – oxidoreductase, sometimes called the cytochrome bc1 complex, and at other times complex III, is the third complex in the electron transport chain, playing a critical role in biochemical generation of ATP. Complex III is a multisubunit transmembrane protein encoded by both the mitochondrial and the nuclear genomes. Complex III is present in the mitochondria of all animals and all aerobic eukaryotes and the inner membranes of most eubacteria. Mutations in Complex III cause exercise intolerance as well as multisystem disorders. The bc1 complex contains 11 subunits, 3 respiratory subunits, 2 core proteins and 6 low-molecular weight proteins.

Coenzyme Q, also known as ubiquinone and marketed as CoQ10, is a coenzyme family that is ubiquitous in animals and most bacteria (hence the name ubiquinone). In humans, the most common form is coenzyme Q10 or ubiquinone-10.
The quinones are a class of organic compounds that are formally "derived from aromatic compounds [such as benzene or naphthalene] by conversion of an even number of –CH= groups into –C(=O)– groups with any necessary rearrangement of double bonds, resulting in "a fully conjugated cyclic dione structure". The archetypical member of the class is 1,4-benzoquinone or cyclohexadienedione, often called simply "quinone". Other important examples are 1,2-benzoquinone (ortho-quinone), 1,4-naphthoquinone and 9,10-anthraquinone.

Succinate dehydrogenase (SDH) or succinate-coenzyme Q reductase (SQR) or respiratory complex II is an enzyme complex, found in many bacterial cells and in the inner mitochondrial membrane of eukaryotes. It is the only enzyme that participates in both the citric acid cycle and the electron transport chain. Histochemical analysis showing high succinate dehydrogenase in muscle demonstrates high mitochondrial content and high oxidative potential.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.
Semiquinone is a free radical resulting from the removal of one hydrogen atom with its electron during the process of dehydrogenation of a hydroquinone, such as hydroquinone itself or catechol, to a quinone or alternatively the addition of a single H atom to a quinone. It is highly unstable.

The cytochrome b6f complex is an enzyme found in the thylakoid membrane in chloroplasts of plants, cyanobacteria, and green algae, that catalyzes the transfer of electrons from plastoquinol to plastocyanin. The reaction is analogous to the reaction catalyzed by cytochrome bc1 of the mitochondrial electron transport chain. During photosynthesis, the cytochrome b6f complex is one step along the chain that transfers electrons from Photosystem II to Photosystem I, and at the same time pumps protons into the thylakoid space, contributing to the generation of an electrochemical (energy) gradient that is later used to synthesize ATP from ADP.

Rieske proteins are iron–sulfur protein (ISP) components of cytochrome bc1 complexes and cytochrome b6f complexes and are responsible for electron transfer in some biological systems. John S. Rieske and co-workers first discovered the protein and in 1964 isolated an acetylated form of the bovine mitochondrial protein. In 1979 Trumpower's lab isolated the "oxidation factor" from bovine mitochondria and showed it was a reconstitutively-active form of the Rieske iron-sulfur protein
It is a unique [2Fe-2S] cluster in that one of the two Fe atoms is coordinated by two histidine residues rather than two cysteine residues. They have since been found in plants, animals, and bacteria with widely ranging electron reduction potentials from -150 to +400 mV.

The Q cycle describes a series of reactions that describe how the sequential oxidation and reduction of the lipophilic electron carrier Coenzyme Q (CoQ) between the ubiquinol and ubiquinone forms, can result in the net movement of protons across a lipid bilayer.

Electron-transferring-flavoprotein dehydrogenase is an enzyme that transfers electrons from electron-transferring flavoprotein in the mitochondrial matrix, to the ubiquinone pool in the inner mitochondrial membrane. It is part of the electron transport chain. The enzyme is found in both prokaryotes and eukaryotes and contains a flavin and FE-S cluster. In humans, it is encoded by the ETFDH gene. Deficiency in ETF dehydrogenase causes the human genetic disease multiple acyl-CoA dehydrogenase deficiency.
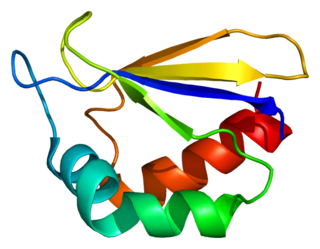
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 is a protein that in humans is encoded by the NDUFA2 gene. The NDUFA2 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Mutations in the NDUFA2 gene are associated with Leigh's syndrome.

Ubiquinol-cytochrome c reductase binding protein, also known as UQCRB, Complex III subunit 7, QP-C, or Ubiquinol-cytochrome c reductase complex 14 kDa protein is a protein which in humans is encoded by the UQCRB gene. This gene encodes a subunit of the ubiquinol-cytochrome c oxidoreductase complex, which consists of one mitochondrial-encoded and 10 nuclear-encoded subunits. Mutations in this gene are associated with mitochondrial complex III deficiency. Alternatively spliced transcript variants have been found for this gene. Related pseudogenes have been identified on chromosomes 1, 5 and X.

NADH:ubiquinone reductase (non-electrogenic) (EC 1.6.5.9, NDH-2, ubiquinone reductase, coenzyme Q reductase, dihydronicotinamide adenine dinucleotide-coenzyme Q reductase, DPNH-coenzyme Q reductase, DPNH-ubiquinone reductase, NADH-coenzyme Q oxidoreductase, NADH-coenzyme Q reductase, NADH-CoQ oxidoreductase, NADH-CoQ reductase) is an enzyme with systematic name NADH:ubiquinone oxidoreductase. This enzyme catalyses the following chemical reaction:

Cytochrome d, previously known as cytochrome a2, is a name for all cytochromes that contain heme D as a cofactor. Two unrelated classes of cytochrome d are known: Cytochrome bd, an enzyme that generates a charge across the membrane so that protons will move, and cytochrome cd1, a nitrite reductase.

Modern biological research has revealed strong evidence that the enzymes of the mitochondrial respiratory chain assemble into larger, supramolecular structures called supercomplexes, instead of the traditional fluid model of discrete enzymes dispersed in the inner mitochondrial membrane. These supercomplexes are functionally active and necessary for forming stable respiratory complexes.

Plácido Navas Lloret is a Spanish Professor of Cell Biology in the Andalusian Center for Developmental Biology at the Pablo de Olavide University in Sevilla, Spain. From 2002 to 2012, Professor Navas served as a board member of the International Coenzyme Q10 Association; since 2013, he has been the chairman of the association.