Defective interfering particles (DIPs), also known as defective interfering viruses, are spontaneously generated virus mutants in which a critical portion of the particle's genome has been lost due to defective replication or non-homologous recombination. The mechanism of their formation is presumed to be as a result of template-switching during replication of the viral genome, although non-replicative mechanisms involving direct ligation of genomic RNA fragments have also been proposed. DIPs are derived from and associated with their parent virus, and particles are classed as DIPs if they are rendered non-infectious due to at least one essential gene of the virus being lost or severely damaged as a result of the defection. A DIP can usually still penetrate host cells, but requires another fully functional virus particle to co-infect a cell with it, in order to provide the lost factors.
An emergent virus is a virus that is either newly appeared, notably increasing in incidence/geographic range or has the potential to increase in the near future. Emergent viruses are a leading cause of emerging infectious diseases and raise public health challenges globally, given their potential to cause outbreaks of disease which can lead to epidemics and pandemics. As well as causing disease, emergent viruses can also have severe economic implications. Recent examples include the SARS-related coronaviruses, which have caused the 2002–2004 outbreak of SARS (SARS-CoV-1) and the 2019–2023 pandemic of COVID-19 (SARS-CoV-2). Other examples include the human immunodeficiency virus, which causes HIV/AIDS; the viruses responsible for Ebola; the H5N1 influenza virus responsible for avian influenza; and H1N1/09, which caused the 2009 swine flu pandemic. Viral emergence in humans is often a consequence of zoonosis, which involves a cross-species jump of a viral disease into humans from other animals. As zoonotic viruses exist in animal reservoirs, they are much more difficult to eradicate and can therefore establish persistent infections in human populations.

Toll-like receptor 7, also known as TLR7, is a protein that in humans is encoded by the TLR7 gene. Orthologs are found in mammals and birds. It is a member of the toll-like receptor (TLR) family and detects single stranded RNA.

Human coronavirus 229E (HCoV-229E) is a species of coronavirus which infects humans and bats. It is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the APN receptor. Along with Human coronavirus OC43, it is one of the viruses responsible for the common cold. HCoV-229E is a member of the genus Alphacoronavirus and subgenus Duvinacovirus.
Spillover infection, also known as pathogen spillover and spillover event, occurs when a reservoir population with a high pathogen prevalence comes into contact with a novel host population. The pathogen is transmitted from the reservoir population and may or may not be transmitted within the host population. Due to climate change and land use expansion, the risk of viral spillover is predicted to significantly increase.

Viral metagenomics uses metagenomic technologies to detect viral genomic material from diverse environmental and clinical samples. Viruses are the most abundant biological entity and are extremely diverse; however, only a small fraction of viruses have been sequenced and only an even smaller fraction have been isolated and cultured. Sequencing viruses can be challenging because viruses lack a universally conserved marker gene so gene-based approaches are limited. Metagenomics can be used to study and analyze unculturable viruses and has been an important tool in understanding viral diversity and abundance and in the discovery of novel viruses. For example, metagenomics methods have been used to describe viruses associated with cancerous tumors and in terrestrial ecosystems.
Katherine Bennett Ensor is an American statistician specializing in numerous methods in computational and statistical analysis of time series data, stochastic process modeling, and estimation to forecast issues in public health, community informatics, computational finance, and environmental statistics.

Tanja Stadler is a mathematician and professor of computational evolution at the Swiss Federal Institute of Technology. She’s the current president of the Swiss Scientific Advisory Panel COVID-19 and Vize-Chair of the Department of Biosystems Science and Engineering at ETH Zürich.

Severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) is a strain of coronavirus that causes COVID-19, the respiratory illness responsible for the COVID-19 pandemic. The virus previously had the provisional name 2019 novel coronavirus (2019-nCoV), and has also been called human coronavirus 2019. First identified in the city of Wuhan, Hubei, China, the World Health Organization designated the outbreak a public health emergency of international concern from January 30, 2020, to May 5, 2023. SARS‑CoV‑2 is a positive-sense single-stranded RNA virus that is contagious in humans.
Pandemic prevention is the organization and management of preventive measures against pandemics. Those include measures to reduce causes of new infectious diseases and measures to prevent outbreaks and epidemics from becoming pandemics.
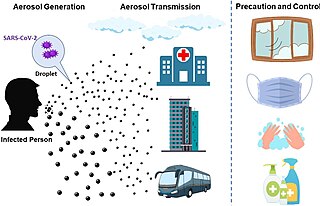
The transmission of COVID-19 is the passing of coronavirus disease 2019 from person to person. COVID-19 is mainly transmitted when people breathe in air contaminated by droplets/aerosols and small airborne particles containing the virus. Infected people exhale those particles as they breathe, talk, cough, sneeze, or sing. Transmission is more likely the closer people are. However, infection can occur over longer distances, particularly indoors.
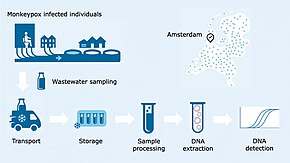
Wastewater-based epidemiology analyzes wastewater to determine the consumption of, or exposure to, chemicals or pathogens in a population. This is achieved by measuring chemical or biomarkers in wastewater generated by the people contributing to a sewage treatment plant catchment. Wastewater-based epidemiology has been used to estimate illicit drug use in communities or populations, but can be used to measure the consumption of alcohol, caffeine, various pharmaceuticals and other compounds. Wastewater-based epidemiology has also been adapted to measure the load of pathogens such as SARS-CoV-2 in a community. It differs from traditional drug testing, urine or stool testing in that results are population-level rather than individual level. Wastewater-based epidemiology is an interdisciplinary endeavour that draws on input from specialists such as wastewater treatment plant operators, analytical chemists and epidemiologists.

The COVID-19 pandemic has affected animals directly and indirectly. SARS-CoV-2, the virus that causes COVID-19, is zoonotic, which likely to have originated from animals such as bats and pangolins. Human impact on wildlife and animal habitats may be causing such spillover events to become much more likely. The largest incident to date was the culling of 14 to 17 million mink in Denmark after it was discovered that they were infected with a mutant strain of the virus.

The COVID-19 Genomics UK (COG-UK) consortium was a group of academic institutions and public health agencies in the United Kingdom created in April 2020 to collect, sequence and analyse genomes of SARS-CoV-2 at scale, as part of COVID-19 pandemic response.

Variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are viruses that, while similar to the original, have genetic changes that are of enough significance to lead virologists to label them separately. SARS-CoV-2 is the virus that causes coronavirus disease 2019 (COVID-19). Some have been stated, to be of particular importance due to their potential for increased transmissibility, increased virulence, or reduced effectiveness of vaccines against them. These variants contribute to the continuation of the COVID-19 pandemic.

A viral vector vaccine is a vaccine that uses a viral vector to deliver genetic material (DNA) that can be transcribed by the recipient's host cells as mRNA coding for a desired protein, or antigen, to elicit an immune response. As of April 2021, six viral vector vaccines, four COVID-19 vaccines and two Ebola vaccines, have been authorized for use in humans.

The nucleocapsid (N) protein is a protein that packages the positive-sense RNA genome of coronaviruses to form ribonucleoprotein structures enclosed within the viral capsid. The N protein is the most highly expressed of the four major coronavirus structural proteins. In addition to its interactions with RNA, N forms protein-protein interactions with the coronavirus membrane protein (M) during the process of viral assembly. N also has additional functions in manipulating the cell cycle of the host cell. The N protein is highly immunogenic and antibodies to N are found in patients recovered from SARS and COVID-19.
ORF10 is an open reading frame (ORF) found in the genome of the SARS-CoV-2 coronavirus. It is 38 codons long. It is not conserved in all Sarbecoviruses. In studies prompted by the COVID-19 pandemic, ORF10 attracted research interest as one of two viral accessory protein genes not conserved between SARS-CoV and SARS-CoV-2 and was initially described as a protein-coding gene likely under positive selection. However, although it is sometimes included in lists of SARS-CoV-2 accessory genes, experimental and bioinformatics evidence suggests ORF10 is likely not a functional protein-coding gene.
ORF1ab refers collectively to two open reading frames (ORFs), ORF1a and ORF1b, that are conserved in the genomes of nidoviruses, a group of viruses that includes coronaviruses. The genes express large polyproteins that undergo proteolysis to form several nonstructural proteins with various functions in the viral life cycle, including proteases and the components of the replicase-transcriptase complex (RTC). Together the two ORFs are sometimes referred to as the replicase gene. They are related by a programmed ribosomal frameshift that allows the ribosome to continue translating past the stop codon at the end of ORF1a, in a -1 reading frame. The resulting polyproteins are known as pp1a and pp1ab.
BA.2.86 is an Omicron subvariant of SARS-CoV-2, the virus that causes COVID-19. BA.2.86 is notable for having more than thirty mutations on its spike protein relative to BA.2. The subvariant, which was first detected in a sample from 24 July 2023, is of concern due to it having made an evolutionary jump on par with the evolutionary jump that the original Omicron variant had made relative to Wuhan-Hu-1, the reference strain first sequenced in Wuhan in December 2019. It is a mutation of BA.2, itself a very early mutation in the Omicron family. BA.2.86 was designated as a variant under monitoring by the World Health Organization on 17 August 2023. The variant was nicknamed Pirola by T. Ryan Gregory, although no official sources use this name. Its descendant JN.1 (BA.2.86.1.1) became the dominating Lineage in Winter 2023/2024.