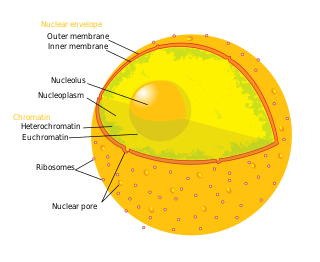
A nuclear pore is a channel as part of the nuclear pore complex (NPC), a large protein complex found in the nuclear envelope in eukaryotic cells, enveloping the cell nucleus containing DNA, which facilitates the selective membrane transport of various molecules across the membrane.

Ran also known as GTP-binding nuclear protein Ran is a protein that in humans is encoded by the RAN gene. Ran is a small 25 kDa protein that is involved in transport into and out of the cell nucleus during interphase and also involved in mitosis. It is a member of the Ras superfamily.
Nuclear transport refers to the mechanisms by which molecules move across the nuclear membrane of a cell. The entry and exit of large molecules from the cell nucleus is tightly controlled by the nuclear pore complexes (NPCs). Although small molecules can enter the nucleus without regulation, macromolecules such as RNA and proteins require association with transport factors known as nuclear transport receptors, like karyopherins called importins to enter the nucleus and exportins to exit.

Drosha is a Class 2 ribonuclease III enzyme that in humans is encoded by the DROSHA gene. It is the primary nuclease that executes the initiation step of miRNA processing in the nucleus. It works closely with DGCR8 and in correlation with Dicer. It has been found significant in clinical knowledge for cancer prognosis and HIV-1 replication.
The HIV-1 Rev response element (RRE) is a highly structured, ~350 nucleotide RNA segment present in the Env coding region of unspliced and partially spliced viral mRNAs. In the presence of the HIV-1 accessory protein Rev, HIV-1 mRNAs that contain the RRE can be exported from the nucleus to the cytoplasm for downstream events such as translation and virion packaging.
Exportin 1 (XPO1), also known as chromosomal region maintenance 1 (CRM1), is a eukaryotic protein that mediates the nuclear export of various proteins and RNAs.

Importin subunit beta-1 is a protein that in humans is encoded by the KPNB1 gene.

RAN binding protein 2 (RANBP2) is protein which in humans is encoded by the RANBP2 gene. It is also known as nucleoporin 358 (Nup358) since it is a member nucleoporin family that makes up the nuclear pore complex. RanBP2 has a mass of 358 kDa.

Nucleoporin 153 (Nup153) is a protein which in humans is encoded by the NUP153 gene. It is an essential component of the basket of nuclear pore complexes (NPCs) in vertebrates, and required for the anchoring of NPCs. It also acts as the docking site of an importing karyopherin. On the cytoplasmic side of the NPC, Nup358 fulfills an analogous role.

Transportin-1 is a protein that in humans is encoded by the TNPO1 gene.
ATP-dependent RNA helicase DDX3X is an enzyme that in humans is encoded by the DDX3X gene.

Ran-specific binding protein 1 is an enzyme that in humans is encoded by the RANBP1 gene.
Ran-binding protein 3 is a protein that in humans is encoded by the RANBP3 gene.

Exportin-T is a protein that in humans is encoded by the XPOT gene.

Exportin-6 is a protein that in humans is encoded by the XPO6 gene.

Importin-13 is a protein encoded by the IPO13 gene in humans. Importin-13 is a member of the importin-β family of nuclear transport receptors (NTRs) and was first identified as a transport receptor in 2000. According to PSI-blast based secondary structure PREDiction (PSIPRED), importin-13 contains 38 α-helices. Importin-13 accommodates a range of cargoes due to its flexible superhelical structure and a cargo binding and release system that is distinct from other importin-like transport receptors. IPO13 is broadly expressed in a variety of tissues in the human body, including the heart, cornea, fetal lung, brain, endometrial carcinoma, and testes.
A nuclear export signal (NES) is a short target peptide containing 4 hydrophobic residues in a protein that targets it for export from the cell nucleus to the cytoplasm through the nuclear pore complex using nuclear transport. It has the opposite effect of a nuclear localization signal, which targets a protein located in the cytoplasm for import to the nucleus. The NES is recognized and bound by exportins.

Rev is a transactivating protein that is essential to the regulation of HIV-1 protein expression. A nuclear localization signal is encoded in the rev gene, which allows the Rev protein to be localized to the nucleus, where it is involved in the export of unspliced and incompletely spliced mRNAs. In the absence of Rev, mRNAs of the HIV-1 late (structural) genes are retained in the nucleus, preventing their translation.
Importin alpha, or karyopherin alpha refers to a class of adaptor proteins that are involved in the import of proteins into the cell nucleus. They are a sub-family of karyopherin proteins.

The microprocessor complex is a protein complex involved in the early stages of processing microRNA (miRNA) and RNA interference (RNAi) in animal cells. The complex is minimally composed of the ribonuclease enzyme Drosha and the dimeric RNA-binding protein DGCR8, and cleaves primary miRNA substrates to pre-miRNA in the cell nucleus. Microprocessor is also the smaller of the two multi-protein complexes that contain human Drosha.