Orthomyxoviridae is a family of negative-sense RNA viruses. It includes seven genera: Alphainfluenzavirus, Betainfluenzavirus, Gammainfluenzavirus, Deltainfluenzavirus, Isavirus, Thogotovirus, and Quaranjavirus. The first four genera contain viruses that cause influenza in birds and mammals, including humans. Isaviruses infect salmon; the thogotoviruses are arboviruses, infecting vertebrates and invertebrates. The Quaranjaviruses are also arboviruses, infecting vertebrates (birds) and invertebrates (arthropods).
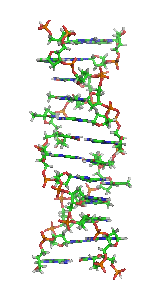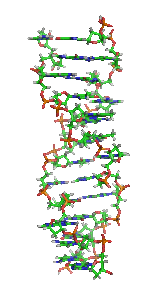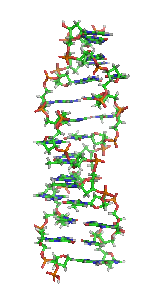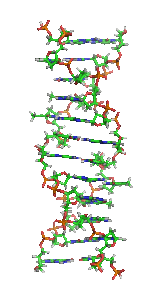
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the helix winds to the left in a zigzag pattern, instead of to the right, like the more common B-DNA form. Z-DNA is thought to be one of three biologically active double-helical structures along with A-DNA and B-DNA.

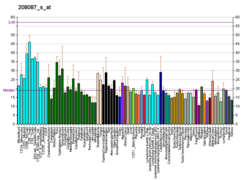
H5N1 genetic structure is the molecular structure of the H5N1 virus's RNA.

Interleukin-1 beta (IL-1β) also known as leukocytic pyrogen, leukocytic endogenous mediator, mononuclear cell factor, lymphocyte activating factor and other names, is a cytokine protein that in humans is encoded by the IL1B gene. There are two genes for interleukin-1 (IL-1): IL-1 alpha and IL-1 beta. IL-1β precursor is cleaved by cytosolic caspase 1 to form mature IL-1β.

Interferon regulatory factor 3, also known as IRF3, is an interferon regulatory factor.

Heterogeneous nuclear ribonucleoprotein A1 is a protein that in humans is encoded by the HNRNPA1 gene. Mutations in hnRNP A1 are causative of amyotrophic lateral sclerosis and the syndrome multisystem proteinopathy.

A virus is a tiny infectious agent that reproduces inside the cells of living hosts. When infected, the host cell is forced to rapidly produce thousands of identical copies of the original virus. Unlike most living things, viruses do not have cells that divide; new viruses assemble in the infected host cell. But unlike simpler infectious agents like prions, they contain genes, which allow them to mutate and evolve. Over 4,800 species of viruses have been described in detail out of the millions in the environment. Their origin is unclear: some may have evolved from plasmids—pieces of DNA that can move between cells—while others may have evolved from bacteria.

The double-stranded RNA-specific adenosine deaminase enzyme family are encoded by the ADAR family genes. ADAR stands for adenosine deaminase acting on RNA. This article focuses on the ADAR proteins; This article details the evolutionary history, structure, function, mechanisms and importance of all proteins within this family.

Mitochondrial antiviral-signaling protein (MAVS) is a protein that is essential for antiviral innate immunity. MAVS is located in the outer membrane of the mitochondria, peroxisomes, and mitochondrial-associated endoplasmic reticulum membrane (MAM). Upon viral infection, a group of cytosolic proteins will detect the presence of the virus and bind to MAVS, thereby activating MAVS. The activation of MAVS leads the virally infected cell to secrete cytokines. This induces an immune response which kills the host's virally infected cells, resulting in clearance of the virus.

Interferon-induced transmembrane protein 1 is a protein that in humans is encoded by the IFITM1 gene. IFITM1 has also recently been designated CD225. This protein has several additional names: fragilis, IFI17 [interferon-induced protein 17], 9-27 [Interferon-inducible protein 9-27] and Leu13.

Cleavage and polyadenylation specificity factor subunit 4 is a protein that in humans is encoded by the CPSF4 gene.

Interferon-induced guanylate-binding protein 2 is a protein that in humans is encoded by the GBP2 gene. GBP2 is a gene related to the superfamily of large GTPases which can be induced mainly by interferon gamma.

Interferon-inducible protein AIM2 also known as absent in melanoma 2 or simply AIM2 is a protein that in humans is encoded by the AIM2 gene.

The nucleotide-binding oligomerization domain-like receptors, or NOD-like receptors (NLRs), are intracellular sensors of pathogen-associated molecular patterns (PAMPs) that enter the cell via phagocytosis or pores, and damage-associated molecular patterns (DAMPs) that are associated with cell stress. They are types of pattern recognition receptors (PRRs), and play key roles in the regulation of innate immune response. NLRs can cooperate with toll-like receptors (TLRs) and regulate inflammatory and apoptotic response.

Influenza, commonly known as "the flu", is an infectious disease caused by influenza viruses. Symptoms range from mild to severe and often include fever, runny nose, sore throat, muscle pain, headache, coughing, and fatigue. These symptoms begin from one to four days after exposure to the virus and last for about 2–8 days. Diarrhea and vomiting can occur, particularly in children. Influenza may progress to pneumonia, which can be caused by the virus or by a subsequent bacterial infection. Other complications of infection include acute respiratory distress syndrome, meningitis, encephalitis, and worsening of pre-existing health problems such as asthma and cardiovascular disease.

In molecular biology, the protein domain Adenosine deaminase z-alpha domain refers to an evolutionary conserved protein domain. This family consists of the N-terminus and thus the z-alpha domain of double-stranded RNA-specific adenosine deaminase (ADAR), an RNA-editing enzyme. The z-alpha domain is a Z-DNA binding domain, and binding of this region to B-DNA has been shown to be disfavoured by steric hindrance.

Stimulator of interferon genes (STING), also known as transmembrane protein 173 (TMEM173) and MPYS/MITA/ERIS is a protein that in humans is encoded by the STING1 gene.
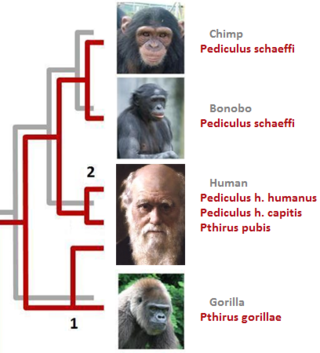
In parasitology and epidemiology, a host switch is an evolutionary change of the host specificity of a parasite or pathogen. For example, the human immunodeficiency virus used to infect and circulate in non-human primates in West-central Africa, but switched to humans in the early 20th century.

Thirumala-Devi Kanneganti is an immunologist and is the Rose Marie Thomas Endowed Chair, Vice Chair of the Department of Immunology, and Member at St. Jude Children's Research Hospital. She is also Director of the Center of Excellence in Innate Immunity and Inflammation at St. Jude Children's Research Hospital. Her research interests include investigating fundamental mechanisms of innate immunity, including inflammasomes and inflammatory cell death, PANoptosis, in infectious and inflammatory disease and cancer.
PANoptosis is an inflammatory cell death pathway. Genetic, molecular, and biochemical studies identified extensive crosstalk among the molecular components across cell death pathways in response to a variety of pathogens and innate immune triggers, leading to the conceptualization of PANoptosis. PANoptosis is defined as a unique innate immune inflammatory cell death pathway driven by caspases and RIPKs and regulated by multi protein PANoptosome complexes. PANoptosis is implicated in driving innate immune responses and inflammation in disease. PANoptosome formation and PANoptosis occur during pathogenic infections, including bacterial, viral, and fungal infections, as well as during inflammatory diseases and can be beneficial in the context of cancer.