Related Research Articles
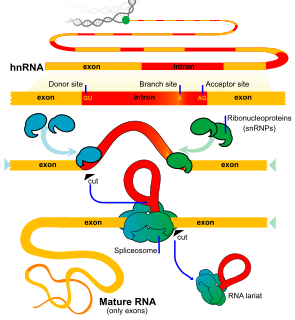
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term exon refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome.
An intron is any nucleotide sequence within a gene that is removed by RNA splicing during maturation of the final RNA product. In other words, introns are non-coding regions of an RNA transcript, or the DNA encoding it, that are eliminated by splicing before translation. The word intron is derived from the term intragenic region, i.e. a region inside a gene. The term intron refers to both the DNA sequence within a gene and the corresponding sequence in RNA transcripts. Sequences that are joined in the final mature RNA after RNA splicing are exons.

RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing all the introns and splicing back together exons. For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, splicing and translation is called gene expression, the central dogma of molecular biology.
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules. Other functions of non-coding DNA include the transcriptional and translational regulation of protein-coding sequences, scaffold attachment regions, origins of DNA replication, centromeres and telomeres. Its RNA counterpart is non-coding RNA.

Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication.

A non-coding RNA (ncRNA) is an RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.
The coding region of a gene, also known as the coding DNA sequence(CDS), is the portion of a gene's DNA or RNA that codes for protein. Studying the length, composition, regulation, splicing, structures, and functions of coding regions compared to non-coding regions over different species and time periods can provide a significant amount of important information regarding gene organization and evolution of prokaryotes and eukaryotes. This can further assist in mapping the human genome and developing gene therapy.

Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.

A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein.

In molecular biology, SNORD116 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

Retinoid X receptor beta (RXR-beta), also known as NR2B2 is a nuclear receptor that in humans is encoded by the RXRB gene.

Spliceosome RNA helicase BAT1 is an enzyme that in humans is encoded by the BAT1 gene.

MHC class II regulatory factor RFX1 is a protein that, in humans, is encoded by the RFX1 gene located on the short arm of chromosome 19.
Splicing factor 1 also known as zinc finger protein 162 (ZFM162) is a protein that in humans is encoded by the SF1 gene.

DNA-directed RNA polymerase I subunit RPA12 is an enzyme that in humans is encoded by the ZNRD1 gene.

TAP2 is a gene in humans that encodes the protein Antigen peptide transporter 2.

RNA-binding protein 39 is a protein that in humans is encoded by the RBM39 gene.

Long non-coding RNAs are a type of RNA, generally defined as transcripts more than 200 nucleotides that are not translated into protein. This arbitrary limit distinguishes long ncRNAs from small non-coding RNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. Long intervening/intergenic noncoding RNAs (lincRNAs) are sequences of lncRNA which do not overlap protein-coding genes.

RING finger protein 39 is a protein that in humans is encoded by the RNF39 gene.

TATA-box binding protein associated factor 7 like is a protein that in humans is encoded by the TAF7L gene.
References
- ↑ Lepourcelet M, Coriton O, Hampe A, Galibert F, Mosser J (1998). "HTEX4, a new human gene in the MHC class I region, undergoes alternative splicing and polyadenylation processes in testis". Immunogenetics. 47 (6): 491–496. doi:10.1007/s002510050388. PMID 9553157. S2CID 6086255.
- ↑ Coriton O, Lepourcelet M, Hampe A, Galibert F, Mosser J (2000). "Transcriptional analysis of the 69-kb sequence centromeric to HLA-J: a dense and complex structure of five genes". Mamm Genome. 11 (12): 1127–1131. doi:10.1007/s003350010213. PMID 11130983. S2CID 22881226.