
A pilus is a hair-like appendage found on the surface of many bacteria and archaea. The terms pilus and fimbria can be used interchangeably, although some researchers reserve the term pilus for the appendage required for bacterial conjugation. All conjugative pili are primarily composed of pilin – fibrous proteins, which are oligomeric.

Escherichia coli ( ESH-ə-RIK-ee-ə KOH-lye) is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes such as EPEC, and ETEC are pathogenic and can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains are part of the normal microbiota of the gut and are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of E. coli benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between E. coli and humans are a type of mutualistic biological relationship — where both the humans and the E. coli are benefitting each other. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.
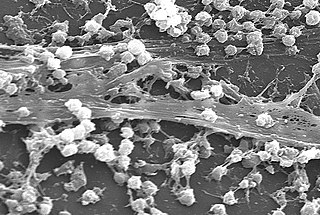
A biofilm is a syntrophic community of microorganisms in which cells stick to each other and often also to a surface. These adherent cells become embedded within a slimy extracellular matrix that is composed of extracellular polymeric substances (EPSs). The cells within the biofilm produce the EPS components, which are typically a polymeric combination of extracellular polysaccharides, proteins, lipids and DNA. Because they have a three-dimensional structure and represent a community lifestyle for microorganisms, they have been metaphorically described as "cities for microbes".

The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis are induced. The system involves the RecA protein. The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (LexA) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.

Filamentation is the anomalous growth of certain bacteria, such as Escherichia coli, in which cells continue to elongate but do not divide. The cells that result from elongation without division have multiple chromosomal copies.
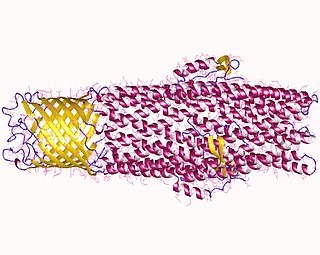
An efflux pump is an active transporter in cells that moves out unwanted material. Efflux pumps are an important component in bacteria in their ability to remove antibiotics. The efflux could also be the movement of heavy metals, organic pollutants, plant-produced compounds, quorum sensing signals, bacterial metabolites and neurotransmitters. All microorganisms, with a few exceptions, have highly conserved DNA sequences in their genome that encode efflux pumps. Efflux pumps actively move substances out of a microorganism, in a process known as active efflux, which is a vital part of xenobiotic metabolism. This active efflux mechanism is responsible for various types of resistance to bacterial pathogens within bacterial species - the most concerning being antibiotic resistance because microorganisms can have adapted efflux pumps to divert toxins out of the cytoplasm and into extracellular media.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.

fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.

RyhB RNA is a 90 nucleotide RNA that down-regulates a set of iron-storage and iron-using proteins when iron is limiting; it is itself negatively regulated by the ferric uptake repressor protein, Fur.
Persister cells are subpopulations of cells that resist treatment, and become antimicrobial tolerant by changing to a state of dormancy or quiescence. Persister cells in their dormancy do not divide. The tolerance shown in persister cells differs from antimicrobial resistance in that the tolerance is not inherited and is reversible. When treatment has stopped the state of dormancy can be reversed and the cells can reactivate and multiply. Most persister cells are bacterial, and there are also fungal persister cells, yeast persister cells, and cancer persister cells that show tolerance for cancer drugs.
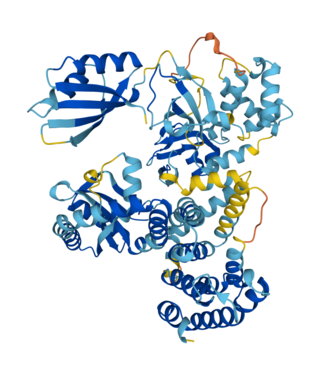
Bifunctional (p)ppGpp synthase/hydrolase SpoT or SpoT is a regulatory enzyme in the RelA/SpoT Homologue (RSH) protein family that synthesizes and hydrolyzes (p)ppGpp to regulate the bacterial stringent response to environmental stressors. SpoT is considered a "long" form RSH protein and is found in many bacteria and plant chloroplasts. SpoT and its homologues have been studied in bacterial model organism E.coli for their role in the production and degradation of (p)ppGpp in the stringent response pathway.
Roberto Kolter is Professor of Microbiology, Emeritus at Harvard Medical School, an author, and past president of the American Society for Microbiology. Kolter has been a professor at Harvard Medical School since 1983 and was Co-director of Harvard's Microbial Sciences Initiative from 2003-2018. During the 35-year term of the Kolter laboratory from 1983 to 2018, more than 130 graduate student and postdoctoral trainees explored an eclectic mix of topics gravitating around the study of microbes. Kolter is a fellow of the American Association for the Advancement of Science and of the American Academy of Microbiology.
Bacterial small RNAs are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.

A toxin-antitoxin system consists of a "toxin" and a corresponding "antitoxin", usually encoded by closely linked genes. The toxin is usually a protein while the antitoxin can be a protein or an RNA. Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies. When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK).
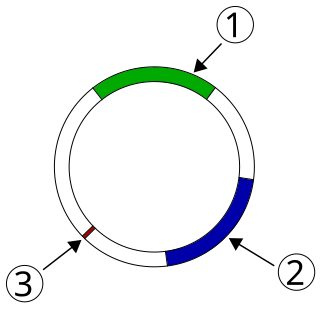
Plasmid-mediated resistance is the transfer of antibiotic resistance genes which are carried on plasmids. Plasmids possess mechanisms that ensure their independent replication as well as those that regulate their replication number and guarantee stable inheritance during cell division. By the conjugation process, they can stimulate lateral transfer between bacteria from various genera and kingdoms. Numerous plasmids contain addiction-inducing systems that are typically based on toxin-antitoxin factors and capable of killing daughter cells that don't inherit the plasmid during cell division. Plasmids often carry multiple antibiotic resistance genes, contributing to the spread of multidrug-resistance (MDR). Antibiotic resistance mediated by MDR plasmids severely limits the treatment options for the infections caused by Gram-negative bacteria, especially family Enterobacteriaceae. The global spread of MDR plasmids has been enhanced by selective pressure from antimicrobial medications used in medical facilities and when raising animals for food.
Thymineless death is the phenomenon by which bacteria, yeasts and mammalian cells undergo cell death when they are starved of thymidine triphosphate (dTTP), an essential precursor for DNA replication. This phenomenon underlies the mechanism of action of several antibacterial, antimalarial and anticancer agents, such as trimethoprim, sulfamethoxazole, methotrexate and fluorouracil.

Escherichia coli is a gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms (endotherms). Most E. coli strains are harmless, but pathogenic varieties cause serious food poisoning, septic shock, meningitis, or urinary tract infections in humans. Unlike normal flora E. coli, the pathogenic varieties produce toxins and other virulence factors that enable them to reside in parts of the body normally not inhabited by E. coli, and to damage host cells. These pathogenic traits are encoded by virulence genes carried only by the pathogens.

The universal stress protein (USP) domain is a superfamily of conserved genes which can be found in bacteria, archaea, fungi, protozoa and plants. Proteins containing the domain are induced by many environmental stressors such as nutrient starvation, drought, extreme temperatures, high salinity, and the presence of uncouplers, antibiotics and metals.
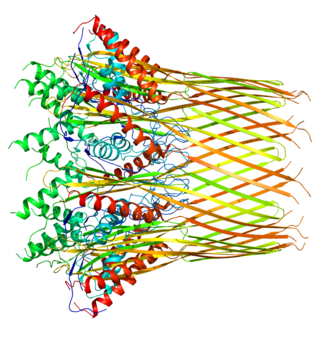
The Curli protein is a type of amyloid fiber produced by certain strains of enterobacteria. They are extracellular fibers located on bacteria such as E. coli and Salmonella spp. These fibers serve to promote cell community behavior through biofilm formation in the extracellular matrix. Amyloids are associated with several human neurodegenerative diseases such as Alzheimer's disease, Huntington's disease, Parkinson's disease, and prion diseases. The study of curli may help to understand human diseases thought to arise from improper amyloid fiber formation. The curli pili are generally assembled through the extracellular nucleation precipitation pathway.

The Phosphate (Pho) regulon is a regulatory mechanism used for the conservation and management of inorganic phosphate within the cell. It was first discovered in Escherichia coli as an operating system for the bacterial strain, and was later identified in other species. The Pho system is composed of various components including extracellular enzymes and transporters that are capable of phosphate assimilation in addition to extracting inorganic phosphate from organic sources. This is an essential process since phosphate plays an important role in cellular membranes, genetic expression, and metabolism within the cell. Under low nutrient availability, the Pho regulon helps the cell survive and thrive despite a depletion of phosphate within the environment. When this occurs, phosphate starvation-inducible (psi) genes activate other proteins that aid in the transport of inorganic phosphate.