
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" base pairs allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins can recognize specific base-pairing patterns that identify particular regulatory regions of genes.

A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet.
Topoisomerases are enzymes that participate in the overwinding or underwinding of DNA. The winding problem of DNA arises due to the intertwined nature of its double-helical structure. During DNA replication and transcription, DNA becomes overwound ahead of a replication fork. If left unabated, this torsion would eventually stop the ability of DNA or RNA polymerases involved in these processes to continue down the DNA strand.
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.

Geminiviridae is a family of plant viruses that encode their genetic information on a circular genome of single-stranded (ss) DNA. There are 520 species in this family, assigned to 14 genera. Diseases associated with this family include: bright yellow mosaic, yellow mosaic, yellow mottle, leaf curling, stunting, streaks, reduced yields. They have single-stranded circular DNA genomes encoding genes that diverge in both directions from a virion strand origin of replication. According to the Baltimore classification they are considered class II viruses. It is the largest known family of single stranded DNA viruses.

A tumour inducing (Ti) plasmid is a plasmid found in pathogenic species of Agrobacterium, including A. tumefaciens, A. rhizogenes, A. rubi and A. vitis.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids. It is widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.

Rolling circle replication (RCR) is a process of unidirectional nucleic acid replication that can rapidly synthesize multiple copies of circular molecules of DNA or RNA, such as plasmids, the genomes of bacteriophages, and the circular RNA genome of viroids. Some eukaryotic viruses also replicate their DNA or RNA via the rolling circle mechanism.
Extrachromosomal DNA is any DNA that is found off the chromosomes, either inside or outside the nucleus of a cell. Most DNA in an individual genome is found in chromosomes contained in the nucleus. Multiple forms of extrachromosomal DNA exist, and, while some of these serve important biological functions, they can also play a role in diseases, such as ecDNA in cancer.

The Argonaute protein family plays a central role in RNA silencing processes, as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the gene silencing phenomenon known as RNA interference (RNAi). Argonaute proteins bind different classes of small non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). Small RNAs guide Argonaute proteins to their specific targets through sequence complementarity, which then leads to mRNA cleavage or translation inhibition.

RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.
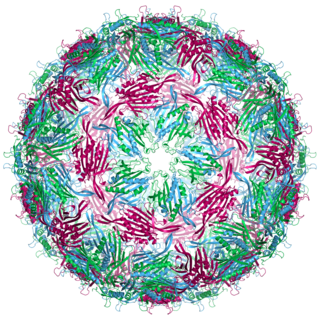
Bacteriophage MS2, commonly called MS2, is an icosahedral, positive-sense single-stranded RNA virus that infects the bacterium Escherichia coli and other members of the Enterobacteriaceae. MS2 is a member of a family of closely related bacterial viruses that includes bacteriophage f2, bacteriophage Qβ, R17, and GA.
PcrA, standing for plasmid copy reduced is a helicase that was originally discovered in a screen for chromosomally encoded genes that are affected in plasmid rolling circle replication in the Gram-positive pathogen Staphylococcus aureus.
In molecular cloning, a vector is a DNA molecule used as a vehicle to artificially carry foreign genetic material into another cell, where it can be replicated and/or expressed. A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids. Common to all engineered vectors have an origin of replication, a multicloning site, and a selectable marker.
Helitrons are one of the three groups of eukaryotic class 2 transposable elements (TEs) so far described. They are the eukaryotic rolling-circle transposable elements which are hypothesized to transpose by a rolling circle replication mechanism via a single-stranded DNA intermediate. They were first discovered in plants and in the nematode Caenorhabditis elegans, and now they have been identified in a diverse range of species, from protists to mammals. Helitrons make up a substantial fraction of many genomes where non-autonomous elements frequently outnumber the putative autonomous partner. Helitrons seem to have a major role in the evolution of host genomes. They frequently capture diverse host genes, some of which can evolve into novel host genes or become essential for Helitron transposition.

Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word cloning refers to the fact that the method involves the replication of one molecule to produce a population of cells with identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA. Molecular cloning methods are central to many contemporary areas of modern biology and medicine.
In cellular biology, the plasmid copy number is the number of copies of a given plasmid in a cell. To ensure survival and thus the continued propagation of the plasmid, they must regulate their copy number. If a plasmid has too high of a copy number, they may excessively burden their host by occupying too much cellular machinery and using too much energy. On the other hand, too low of a copy number may result in the plasmid not being present in all of their host's progeny. Plasmids may be either low, medium or high copy number plasmids; the regulation mechanisms between low and medium copy number plasmids are different. Low copy plasmids require either a partitioning system or a toxin-antitoxin pair such as CcdA/CcdB to ensure that each daughter receives the plasmid. For example, the F plasmid, which is the origin of BACs is a single copy plasmid with a partitioning system encoded in an operon right next to the plasmid origin. The partitioning system interacts with the septation apparatus to ensure that each daughter receives a copy of the plasmid. Many biotechnology applications utilize mutated plasmids that replicate to high copy number. For example, pBR322 is a medium copy number plasmid from which several high copy number cloning vectors have been derived by mutagenesis, such as the well known pUC series. This delivers the convenience of high plasmid DNA yields but the additional burden of the high copy number restricts the plasmid size. Larger high copy plasmids (>30kb) are disfavoured and also prone to size reduction through deletional mutagenesis.

The GC-Enriched, Between Replication Origins RNA motif is a conserved RNA or single-stranded DNA structure that was discovered by bioinformatics. Although the GEBRO motif was published as an RNA candidate, there is some reason to suspect that it might function as a single-stranded DNA. In terms of secondary structure, RNA and DNA are difficult to distinguish when only sequence information is available. GEBRO motifs are found in some species of Streptococcus.
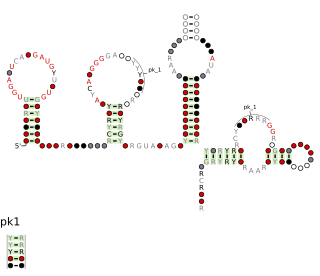
The Plasmid-Associated gamma-Proteobacteria Especially Vibrionales RNA motif is a conserved RNA structure that was discovered by bioinformatics. PAGEV motif RNAs are found in Gammaproteobacteria, especially within the order Vibrionales.

Monodnaviria is a realm of viruses that includes all single-stranded DNA viruses that encode an endonuclease of the HUH superfamily that initiates rolling circle replication of the circular viral genome. Viruses descended from such viruses are also included in the realm, including certain linear single-stranded DNA (ssDNA) viruses and circular double-stranded DNA (dsDNA) viruses. These atypical members typically replicate through means other than rolling circle replication.













