
In molecular biology, Small Cajal body specific RNA 14 is a small nucleolar RNA found in Cajal bodies.

In molecular biology, snoRNA U101 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, snoRNA U102 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA U103 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, snoRNA U95 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, the small nucleolar RNAs SNORD106 and SNORD12 are two related snoRNAs which belongs to the C/D class of small nucleolar RNAs (snoRNAs). Both contain the conserved C (UGAUGA) and D (CUGA) box sequence motifs

DEAD box proteins are involved in an assortment of metabolic processes that typically involve RNAs, but in some cases also other nucleic acids. They are highly conserved in nine motifs and can be found in most prokaryotes and eukaryotes, but not all. Many organisms, including humans, contain DEAD-box (SF2) helicases, which are involved in RNA metabolism.

Tetraloops are a type of four-base hairpin loop motifs in RNA secondary structure that cap many double helices. There are many variants of the tetraloop. The published ones include ANYA, CUYG, GNRA, UNAC and UNCG.

The Moco RNA motif is a conserved RNA structure that is presumed to be a riboswitch that binds molybdenum cofactor or the related tungsten cofactor. Genetic experiments support the hypothesis that the Moco RNA motif corresponds to a genetic control element that responds to changing concentrations of molybdenum or tungsten cofactor. As these cofactors are not available in purified form, in vitro binding assays cannot be performed. However, the genetic data, complex structure of the RNA and the failure to detect a protein involved in the regulation suggest that the Moco RNA motif corresponds to a class of riboswitches.

6C RNA is a class of non-coding RNA present in actinomycetes. 6C RNA was originally discovered as a conserved RNA structure having two stem-loops each containing six or more cytosine (C) residues. Later work revealed that 6C RNAs in Streptomyces coelicolor and Streptomyces avermitilis have predicted rho-independent transcription terminators, and microarray and reverse-transcriptase PCR experiments indicate that the S. coelicolor version is transcribed as RNA. Transcription of the S. coelicolor RNA increases during sporulation, and three transcripts were detected that overlap the 6C motif, but have different apparent start and stop sites.
Internal-loops in RNA are found where the double stranded RNA separates due to no Watson-Crick-Franklin base pairing between the nucleotides. Internal-loops differ from Stem-loops as they occur in middle of a stretch of double stranded RNA. The non-canonicoal residues result in the double helix becoming distorted due to unwinding, unstacking and kinking.

RNAs Associated with Genes Associated with Twister and Hammerhead ribozymes (RAGATH) refers to a bioinformatics strategy that was devised to find self-cleaving ribozymes in bacteria. It also refers to candidate RNAs, or RAGATH RNA motifs, discovered using this strategy.
In archaea like in eukaryotes, uridines in various RNAs are converted to pseudouridines by ribonucleoprotein complexes (RNPs) containing H/ACA sRNA. Because of their conserved function, these sRNAs are also called small "nucleolar" RNAs (snoRNA) like in eukaryotes, despite no nucleus is present in prokaryotes. By using various computational and experimental approaches in three Pyrococcus genomes seven H/ACA sRNAs and 15 pseudouridine (Ψ) resides on rRNA were identified. One H/ACA motif was shown to guide up to three distinct pseudouridylations. Atypical pseudouridine guide RNA features were identified in Pyrobaculum species. Lack of the conserved 3'-terminal ACA sequence and sometimes lack of 5' portion of the pseudouridylation pocket feature in few conserved Pyrobaculum H/ACA-like sRNAs. A study by Toffano-Nioche et al. proposes an unified structure/function model based on the common structural components in "Euryarchaeota" and Thermoproteota shared by H/ACA and H/ACA-like motifs.
Non-coding RNAs have been discovered using both experimental and bioinformatic approaches. Bioinformatic approaches can be divided into three main categories. The first involves homology search, although these techniques are by definition unable to find new classes of ncRNAs. The second category includes algorithms designed to discover specific types of ncRNAs that have similar properties. Finally, some discovery methods are based on very general properties of RNA, and are thus able to discover entirely new kinds of ncRNAs.
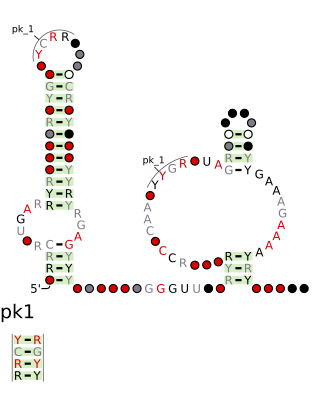
The drum RNA motif is a conserved RNA structure that was discovered by bioinformatics. Drum motifs are found in Bacillota, Bacteroidota, Pseudomonadota, and Spirochaetota, and exhibit multiple highly conserved nucleotide positions, despite their widespread distribution.

The DUF3268 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF3268 motifs are found in Bacillota and Clostridia.

The FuFi-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Such RNA "motifs" are often the first step to elucidating the biological function of a novel RNA. FuFi-1 motif RNAs are found in Bacillota AND Fusobacteriota.

The Human, Oral, Large, Distant to HINT RNA motif is a conserved RNA structure that was discovered by bioinformatics. HOLDH motif RNAs are found exclusively in metagenomic sequences, especially those derived from human supragingival dental plaque and on the tongue.
An RNA motif is a description of a group of RNAs that have a related structure. RNA motifs consist of a pattern of features within the primary sequence and secondary structure of related RNAs. Thus, it extends the concept of a sequence motif to include RNA secondary structure. The term "RNA motif" can refer both to the pattern and to the RNA sequences that match it.















