Related Research Articles

Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass.
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.
A release factor is a protein that allows for the termination of translation by recognizing the termination codon or stop codon in an mRNA sequence. They are named so because they release new peptides from the ribosome.

Eukaryotic translation termination factor 1 (eRF1), also known as TB3-1, is a protein that in humans is encoded by the ETF1 gene.

50S is the larger subunit of the 70S ribosome of prokaryotes, i.e. bacteria and archaea. It is the site of inhibition for antibiotics such as macrolides, chloramphenicol, clindamycin, and the pleuromutilins. It includes the 5S ribosomal RNA and 23S ribosomal RNA.

The 23S rRNA is a 2,904 nucleotide long component of the large subunit (50S) of the bacterial/archean ribosome and makes up the peptidyl transferase center (PTC). The 23S rRNA is divided into six secondary structural domains titled I-VI, with the corresponding 5S rRNA being considered domain VII. The ribosomal peptidyl transferase activity resides in domain V of this rRNA, which is also the most common binding site for antibiotics that inhibit translation, making it a target for ribosomal engineering. A well-known member of this antibiotic class, chloramphenicol, acts by inhibiting peptide bond formation, with recent 3D-structural studies showing two different binding sites depending on the species of ribosome. Numerous mutations in domains of the 23S rRNA with Peptidyl transferase activity have resulted in antibiotic resistance. 23S rRNA genes typically have higher sequence variations, including insertions and/or deletions, compared to other rRNAs.

Splicing factor U2AF 35 kDa subunit is a protein that in humans is encoded by the U2AF1 gene.

Dimethyladenosine transferase 1, mitochondrial; Transcription factor B1, mitochondrial is a mitochondrial enzyme that in is encoded by the TFB1M gene.
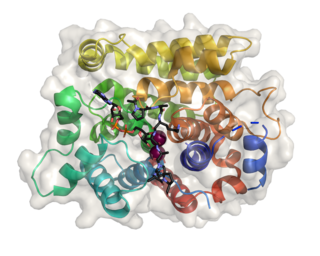
(ADP-ribosyl)hydrolase 3 (ARH3) is an enzyme that in humans is encoded by the ADPRHL2 gene (also called ADPRS). This enzyme reverses the proteins’ post-translational addition of ADP-ribose to serine residues as part of the DNA damage response The enzyme is also known to cleave poly(ADP-ribose) polymers, 1''-O-acetyl-ADP-ribose and alpha-NAD+

EF-G is a prokaryotic elongation factor involved in protein translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.

Mitochondrial translational release factor 1, also known as MTRF1 is a human gene.
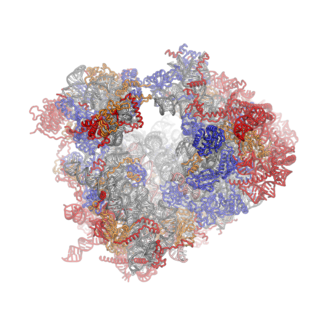
Ribosomes are a large and complex molecular machine that catalyzes the synthesis of proteins, referred to as translation. The ribosome selects aminoacylated transfer RNAs (tRNAs) based on the sequence of a protein-encoding messenger RNA (mRNA) and covalently links the amino acids into a polypeptide chain. Ribosomes from all organisms share a highly conserved catalytic center. However, the ribosomes of eukaryotes are much larger than prokaryotic ribosomes and subject to more complex regulation and biogenesis pathways. Eukaryotic ribosomes are also known as 80S ribosomes, referring to their sedimentation coefficients in Svedberg units, because they sediment faster than the prokaryotic (70S) ribosomes. Eukaryotic ribosomes have two unequal subunits, designated small subunit (40S) and large subunit (60S) according to their sedimentation coefficients. Both subunits contain dozens of ribosomal proteins arranged on a scaffold composed of ribosomal RNA (rRNA). The small subunit monitors the complementarity between tRNA anticodon and mRNA, while the large subunit catalyzes peptide bond formation.
Diadenosine tetraphosphate or Ap4A is a putative alarmone, ubiquitous in nature being common to everything from bacteria to humans. It is made up of two adenosines joined together by a 5′-5′ linked chain of four phosphates. Adenosine polyphosphates are capable of inducing multiple physiological effects.

EF-P is an essential protein that in bacteria stimulates the formation of the first peptide bonds in protein synthesis. Studies show that EF-P prevents ribosomes from stalling during the synthesis of proteins containing consecutive prolines. EF-P binds to a site located between the binding site for the peptidyl tRNA and the exiting tRNA. It spans both ribosomal subunits with its amino-terminal domain positioned adjacent to the aminoacyl acceptor stem and its carboxyl-terminal domain positioned next to the anticodon stem-loop of the P site-bound initiator tRNA. The EF-P protein shape and size is very similar to a tRNA and interacts with the ribosome via the exit “E” site on the 30S subunit and the peptidyl-transferase center (PTC) of the 50S subunit. EF-P is a translation aspect of an unknown function, therefore It probably functions indirectly by altering the affinity of the ribosome for aminoacyl-tRNA, thus increasing their reactivity as acceptors for peptidyl transferase.
Susan L. Ackerman is an American neuroscientist and geneticist. Her work has highlighted some of the genetic and biochemical factors that are involved in the development of the central nervous system and age-related neurodegeneration. Her research is aimed at helping scientists understand what causes several types of neurodegeneration in mammals. This research, and others' like it, may lead to cures for neurodegenerative diseases. Ackerman is a professor at University of California San Diego. She was formerly a professor at the Jackson Laboratory and the Sackler School of Graduate Biomedical Sciences at Tufts University. She also serves as an adjunct professor at the University of Maine, Orono. Ackerman was an associate geneticist at Massachusetts General Hospital in Boston, Massachusetts.

Ribosomal pause refers to the queueing or stacking of ribosomes during translation of the nucleotide sequence of mRNA transcripts. These transcripts are decoded and converted into an amino acid sequence during protein synthesis by ribosomes. Due to the pause sites of some mRNA's, there is a disturbance caused in translation. Ribosomal pausing occurs in both eukaryotes and prokaryotes. A more severe pause is known as a ribosomal stall.
Alternative ribosome-rescue factor A also known as peptidyl-tRNA hydrolase, is a protein that plays a role in rescuing of stalled ribosomes. It recruits RF2.

Zinc finger protein 598 (ZNF598) is a protein that in humans is encoded by the ZNF598 gene.
ORF10 is an open reading frame (ORF) found in the genome of the SARS-CoV-2 coronavirus. It is 38 codons long. It is not conserved in all Sarbecoviruses. In studies prompted by the COVID-19 pandemic, ORF10 attracted research interest as one of two viral accessory protein genes not conserved between SARS-CoV and SARS-CoV-2 and was initially described as a protein-coding gene likely under positive selection. However, although it is sometimes included in lists of SARS-CoV-2 accessory genes, experimental and bioinformatics evidence suggests ORF10 is likely not a functional protein-coding gene.
References
- 1 2 "Peptidyl-tRNA hydrolase, ribosome rescue factor". BioCyc. SRI International.
- ↑ Chan KH, Petrychenko V, Mueller C, Maracci C, Holtkamp W, Wilson DN, et al. (August 2020). "Mechanism of ribosome rescue by alternative ribosome-rescue factor B". Nature Communications. 11 (1): 4106. Bibcode:2020NatCo..11.4106C. doi:10.1038/s41467-020-17853-7. PMC 7427801 . PMID 32796827.
- ↑ Carbone CE, Demo G, Madireddy R, Svidritskiy E, Korostelev AA (November 2020). "ArfB can displace mRNA to rescue stalled ribosomes". Nature Communications. 11 (1): 5552. Bibcode:2020NatCo..11.5552C. doi:10.1038/s41467-020-19370-z. PMC 7641280 . PMID 33144582.
- ↑ Feaga HA, Quickel MD, Hankey-Giblin PA, Keiler KC (March 2016). "Human Cells Require Non-stop Ribosome Rescue Activity in Mitochondria". PLOS Genetics. 12 (3): e1005964. doi:10.1371/journal.pgen.1005964. PMC 4814080 . PMID 27029019.
- ↑ Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, et al. (March 2010). "A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome". The EMBO Journal. 29 (6): 1116–1125. doi:10.1038/emboj.2010.14. PMC 2845271 . PMID 20186120.