
In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome.

A basic helix–loop–helix (bHLH) is a protein structural motif that characterizes one of the largest families of dimerizing transcription factors. The word "basic" does not refer to complexity but to the chemistry of the motif because transcription factors in general contain basic amino acid residues in order to facilitate DNA binding.
Cis-regulatory elements (CREs) or Cis-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology.

Apetala 2(AP2) is a gene and a member of a large family of transcription factors, the AP2/EREBP family. In Arabidopsis thaliana AP2 plays a role in the ABC model of flower development. It was originally thought that this family of proteins was plant-specific; however, recent studies have shown that apicomplexans, including the causative agent of malaria, Plasmodium falciparum encode a related set of transcription factors, called the ApiAP2 family.

Activating transcription factor 6, also known as ATF6, is a protein that, in humans, is encoded by the ATF6 gene and is involved in the unfolded protein response.

D site of albumin promoter binding protein, also known as DBP, is a protein which in humans is encoded by the DBP gene.
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global binding sites precisely for any protein of interest. Previously, ChIP-on-chip was the most common technique utilized to study these protein–DNA relations.
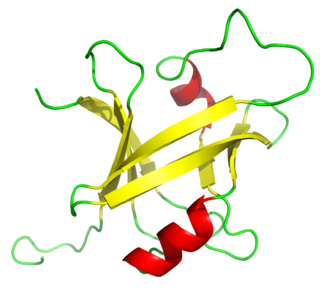
The B3 DNA binding domain (DBD) is a highly conserved domain found exclusively in transcription factors combined with other domains. It consists of 100-120 residues, includes seven beta strands and two alpha helices that form a DNA-binding pseudobarrel protein fold ; it interacts with the major groove of DNA.

A biological network is a method of representing systems as complex sets of binary interactions or relations between various biological entities. In general, networks or graphs are used to capture relationships between entities or objects. A typical graphing representation consists of a set of nodes connected by edges.
CADgene is a database of genes involved in coronary artery disease (CAD).

The Basic Leucine Zipper Domain is found in many DNA binding eukaryotic proteins. One part of the domain contains a region that mediates sequence specific DNA binding properties and the leucine zipper that is required to hold together (dimerize) two DNA binding regions. The DNA binding region comprises a number of basic amino acids such as arginine and lysine. Proteins containing this domain are transcription factors.
The Functional Element SNPs Database (FESD) is a biological database of single nucleotide polymorphisms in molecular biology. The database is a tool designed to organize functional elements into categories in human gene regions and to output their sequences needed for genotyping experiments as well as provide a set of SNPs that lie within each region. The database defines functional elements into ten types: promoter regions, CpG islands,5' untranslated regions (5'-UTRs), translation start sites, splice sites, coding exons, introns, translation stop sites, polyadenylation signals, and 3' UTRs. People may reference this database for haplotype information or obtain a flanking sequence for genotyping. This may help in finding mutations that contribute to common and polygenic diseases. Researchers can manually choose a group of SNPs of special interest for certain functional elements along with their corresponding sequences. The database combines information from sources such as HapMap, UCSC GoldenPath, dbSNP, OMIM, and TRANSFAC. Users can obtain information about tag SNPs and simulate LD blocks for each gene. FESD is still a developing database and is not widely known so was unable to find projects that used the database. Research was found using similar databases or databases that are combined in FESD’s information pool.

The SQUAMOSA promoter binding protein-like family of transcription factors are defined by a plant-specific DNA-binding domain. The founding member of the family was identified based on its specific in vitro binding to the promoter of the snapdragon SQUAMOSA gene. SBP proteins are thought to be transcriptional activators.
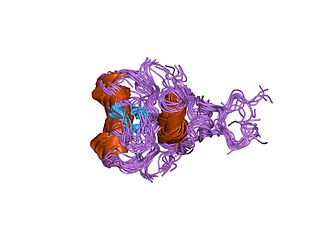
In molecular biology, GATA zinc fingers are zinc-containing domains found in a number of transcription factors. Some members of this class of zinc fingers specifically bind the DNA sequence (A/T)GATA(A/G) in the regulatory regions of genes., giving rise to the name of the domain. In these domains, a single zinc ion is coordinated by 4 cysteine residues. NMR studies have shown the core of the Znf to comprise 2 irregular anti-parallel beta-sheets and an alpha-helix, followed by a long loop to the C-terminal end of the finger. The N-terminal part, which includes the helix, is similar in structure, but not sequence, to the N-terminal zinc module of the glucocorticoid receptor DNA-binding domain. The helix and the loop connecting the 2 beta-sheets interact with the major groove of the DNA, while the C-terminal tail wraps around into the minor groove. Interactions between the Znf and DNA are mainly hydrophobic, explaining the preponderance of thymines in the binding site; a large number of interactions with the phosphate backbone have also been observed. Two GATA zinc fingers are found in GATA-family transcription factors. However, there are several proteins that only contains a single copy of the domain. It is also worth noting that many GATA-type Znfs have not been experimentally demonstrated to be DNA-binding domains. Furthermore, several GATA-type Znfs have been demonstrated to act as protein-recognition domains. For example, the N-terminal Znf of GATA1 binds specifically to a zinc finger from the transcriptional coregulator FOG1 (ZFPM1).

The WRKY domain is found in the WRKY transcription factor family, a class of transcription factors. The WRKY domain is found almost exclusively in plants although WRKY genes appear present in some diplomonads, social amoebae and other amoebozoa, and fungi incertae sedis. They appear absent in other non-plant species. WRKY transcription factors have been a significant area of plant research for the past 20 years. The WRKY DNA-binding domain recognizes the W-box (T)TGAC(C/T) cis-regulatory element.
In molecular biology, the protein domain TCP is actually a family of transcription factors named after: teosinte branched 1, cycloidea (cyc) and PCF in rice.
In molecular biology the protein domain Whirly is a transcription factor commonly found in plants. This means they aid the transcription of genes from DNA into a complementary copy of mRNA. In particular, in plants, they aid the transcription of plant defence genes.

Myelin regulatory factor, also known as myelin gene regulatory factor (MRF), is a protein that in humans is encoded by the MYRF gene.
Identification of genomic regulatory elements is essential for understanding the dynamics of developmental, physiological and pathological processes. Recent advances in chromatin immunoprecipitation followed by sequencing (ChIP-seq) have provided powerful ways to identify genome-wide profiling of DNA-binding proteins and histone modifications. The application of ChIP-seq methods has reliably discovered transcription factor binding sites and histone modification sites.
JASPAR is an open access and widely used database of manually curated, non-redundant transcription factor (TF) binding profiles stored as position frequency matrices (PFM) and transcription factor flexible models (TFFM) for TFs from species in six taxonomic groups. From the supplied PFMs, users may generate position-specific weight matrices (PWM). The JASPAR database was introduced in 2004. There were seven major updates and new releases in 2006, 2008, 2010, 2014, 2016, 2018, 2020 and 2022, which is the latest release of JASPAR.











