The 5′ untranslated region is the region of an mRNA that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming complex secondary structure to regulate translation. The 5′ UTR has been found to interact with proteins relating to metabolism; and proteins translate sequences within the 5′ UTR. In addition, this region has been involved in transcription regulation, such as the sex-lethal gene in Drosophila. Regulatory elements within 5′ UTRs have also been linked to mRNA export.
An internal ribosome entry site, abbreviated IRES, is an RNA element that allows for translation initiation in a cap-independent manner, as part of the greater process of protein synthesis. In eukaryotic translation, initiation typically occurs at the 5' end of mRNA molecules, since 5' cap recognition is required for the assembly of the initiation complex. The location for IRES elements is often in the 5'UTR, but can also occur elsewhere in mRNAs.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recycling.
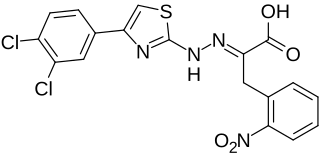
4EGI-1 is a synthetic chemical compound which has been found to interfere with the growth of certain types of cancer cells in vitro. Its mechanism of action involves interruption of the binding of cellular initiation factor proteins involved in the translation of transcribed mRNA at the ribosome. The inhibition of these initiation factors prevents the initiation and translation of many proteins whose functions are essential to the rapid growth and proliferation of cancer cells.

The BiP internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of BiP protein and allows cap-independent translation. BiP protein expression has been found to be significantly enhanced by the heat shock response due to internal ribosome entry site (IRES)-dependent translation. It is thought that this translational mechanism is essential for the survival of cells under stress.
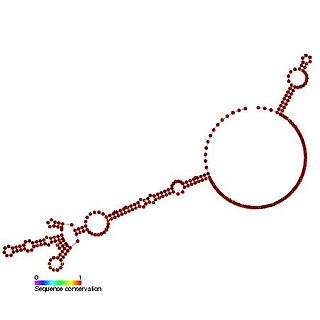
The c-myc internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of C-myc and allows cap-independent translation. The mammalian c-myc gene is a proto-oncogene which is required for cell proliferation, transformation and death. c-myc mRNA has an alternative method of translation via internal ribosome entry where ribosomes are recruited to the IRES located in the 5' UTR thus bypassing the typical eukaryotic cap-dependent translation pathway.
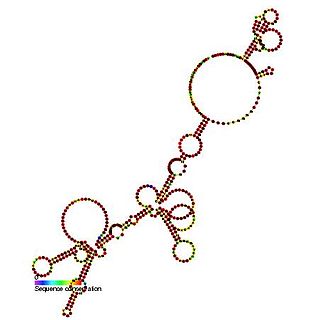
The FGF-2 internal ribosome entry site is an RNA element present in the 5' UTR of the mRNA of fibroblast growth factor-2. It has been found that the FGF-2 internal ribosome entry site (IRES) activity is strictly controlled and highly tissue specific. It is thought that translational IRES dependent activation of FGF-2 plays a vital role in embryogenesis and in the adult brain [1]. When expressed the fibroblast growth factor 2 FGF-2 protein plays a pivotal role in cell proliferation, differentiation and survival as well as being involved in wound-healing [1,2].
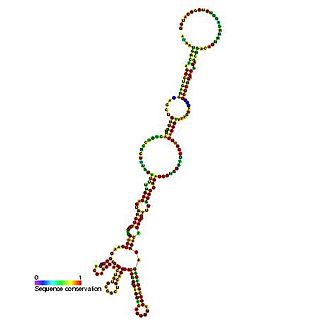
The heat shock protein 70 (Hsp70) internal ribosome entry site (IRES) is an RNA element that allows cap independent translation during conditions such as heat shock and stress. It has been shown that the 216 nucleotide long 5' UTR contains internal ribosome entry site activity.

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.
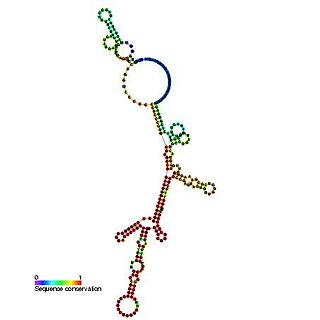
The Hepatitis C virus internal ribosome entry site, or HCV IRES, is an RNA structure within the 5'UTR of the HCV genome that mediates cap-independent translation initiation.

The L-myc internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of L-myc that allows cap-independent translation. L-myc undergoes translation via the internal ribosome entry site and bypasses the typical eukaryotic cap-dependent translation pathway [1]. The myc family of genes when expressed are known to be involved in the control of cell growth, differentiation and apoptosis.

The Mnt internal ribosome entry site (IRES) is an RNA element. Mnt is a transcriptional repressor related to the Myc/Mad family of transcription factors. It is thought that this IRES allows efficient Mnt synthesis when cap-dependent translation initiation is reduced.

The N-myc internal ribosome entry site (IRES) is an RNA element found in the n-myc gene. The myc family of genes when expressed are known to be involved in the control of cell growth, differentiation and apoptosis. n-myc mRNA has an alternative method of translation via an internal ribosome entry site where ribosomes are recruited to the IRES located in the 5' UTR thus bypassing the typical eukaryotic cap-dependent translation pathway.

The HIF-1α internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of HIF-1α that allows cap-independent translation. The HIF-1α internal ribosome entry site (IRES) allows translation to be maintained under hypoxic cell conditions that inhibit cap-dependent translation [1]. The hypoxia-inducible factor-1α protein (HIF-1α) is a subunit of the HIF-1 transcription factor, which induces transcription of several genes involved in the cellular response to hypoxia.
A ribosome binding site, or ribosomal binding site (RBS), is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of protein translation. Mostly, RBS refers to bacterial sequences, although internal ribosome entry sites (IRES) have been described in mRNAs of eukaryotic cells or viruses that infect eukaryotes. Ribosome recruitment in eukaryotes is generally mediated by the 5' cap present on eukaryotic mRNAs.
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human eIF4G 1 has been the focus of extensive studies.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3′ and 5′ untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5′ cap is attached onto mRNA and this 5′ cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3′ UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.
Translational regulation refers to the control of the levels of protein synthesized from its mRNA. This regulation is vastly important to the cellular response to stressors, growth cues, and differentiation. In comparison to transcriptional regulation, it results in much more immediate cellular adjustment through direct regulation of protein concentration. The corresponding mechanisms are primarily targeted on the control of ribosome recruitment on the initiation codon, but can also involve modulation of peptide elongation, termination of protein synthesis, or ribosome biogenesis. While these general concepts are widely conserved, some of the finer details in this sort of regulation have been proven to differ between prokaryotic and eukaryotic organisms.
In molecular biology, the ODC internal ribosome entry site (IRES) is an RNA element present in the 5′ UTR of the mRNA encoding ornithine decarboxylase. It has been suggested that this IRES allows cap-independent translation of ornithine decarboxylase at the G2/M phase of the cell cycle, however there is some doubt about this. Translation from this IRES is activated by the zinc finger protein ZNF9 and by Poly(rC)-binding protein 2 (PCBP2). It is also activated in Ras-transformed cells.












