The Semantic Web, sometimes known as Web 3.0, is an extension of the World Wide Web through standards set by the World Wide Web Consortium (W3C). The goal of the Semantic Web is to make Internet data machine-readable.
The Resource Description Framework (RDF) is a World Wide Web Consortium (W3C) standard originally designed as a data model for metadata. It has come to be used as a general method for description and exchange of graph data. RDF provides a variety of syntax notations and data serialization formats, with Turtle currently being the most widely used notation.
The Web Ontology Language (OWL) is a family of knowledge representation languages for authoring ontologies. Ontologies are a formal way to describe taxonomies and classification networks, essentially defining the structure of knowledge for various domains: the nouns representing classes of objects and the verbs representing relations between the objects.
SPARQL is an RDF query language—that is, a semantic query language for databases—able to retrieve and manipulate data stored in Resource Description Framework (RDF) format. It was made a standard by the RDF Data Access Working Group (DAWG) of the World Wide Web Consortium, and is recognized as one of the key technologies of the semantic web. On 15 January 2008, SPARQL 1.0 was acknowledged by W3C as an official recommendation, and SPARQL 1.1 in March, 2013.

FOAF is a machine-readable ontology describing persons, their activities and their relations to other people and objects. Anyone can use FOAF to describe themselves. FOAF allows groups of people to describe social networks without the need for a centralised database.
Notation3, or N3 as it is more commonly known, is a shorthand non-XML serialization of Resource Description Framework models, designed with human-readability in mind: N3 is much more compact and readable than XML RDF notation. The format is being developed by Tim Berners-Lee and others from the Semantic Web community. A formalization of the logic underlying N3 was published by Berners-Lee and others in 2008.
RDFa or Resource Description Framework in Attributes is a W3C Recommendation that adds a set of attribute-level extensions to HTML, XHTML and various XML-based document types for embedding rich metadata within Web documents. The Resource Description Framework (RDF) data-model mapping enables its use for embedding RDF subject-predicate-object expressions within XHTML documents. It also enables the extraction of RDF model triples by compliant user agents.
In computing, Terse RDF Triple Language (Turtle) is a syntax and file format for expressing data in the Resource Description Framework (RDF) data model. Turtle syntax is similar to that of SPARQL, an RDF query language. It is a common data format for storing RDF data, along with N-Triples, JSON-LD and RDF/XML.
An RDF query language is a computer language, specifically a query language for databases, able to retrieve and manipulate data stored in Resource Description Framework (RDF) format.
Semantic publishing on the Web, or semantic web publishing, refers to publishing information on the web as documents accompanied by semantic markup. Semantic publication provides a way for computers to understand the structure and even the meaning of the published information, making information search and data integration more efficient.
Semantically Interlinked Online Communities Project is a Semantic Web technology. SIOC provides methods for interconnecting discussion methods such as blogs, forums and mailing lists to each other. It consists of the SIOC ontology, an open-standard machine-readable format for expressing the information contained both explicitly and implicitly in Internet discussion methods, of SIOC metadata producers for a number of popular blogging platforms and content management systems, and of storage and browsing/searching systems for leveraging this SIOC data.
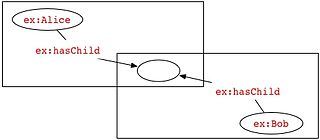
In RDF, a blank node is a node in an RDF graph representing a resource for which a URI or literal is not given. The resource represented by a blank node is also called an anonymous resource. According to the RDF standard a blank node can only be used as subject or object of an RDF triple.

In computing, linked data is structured data which is interlinked with other data so it becomes more useful through semantic queries. It builds upon standard Web technologies such as HTTP, RDF and URIs, but rather than using them to serve web pages only for human readers, it extends them to share information in a way that can be read automatically by computers. Part of the vision of linked data is for the Internet to become a global database.

DBpedia is a project aiming to extract structured content from the information created in the Wikipedia project. This structured information is made available on the World Wide Web. DBpedia allows users to semantically query relationships and properties of Wikipedia resources, including links to other related datasets.
N-Triples is a format for storing and transmitting data. It is a line-based, plain text serialisation format for RDF graphs, and a subset of the Turtle format. N-Triples should not be confused with Notation3 which is a superset of Turtle. N-Triples was primarily developed by Dave Beckett at the University of Bristol and Art Barstow at the World Wide Web Consortium (W3C).

In computer science, information science and systems engineering, ontology engineering is a field which studies the methods and methodologies for building ontologies, which encompasses a representation, formal naming and definition of the categories, properties and relations between the concepts, data and entities. In a broader sense, this field also includes a knowledge construction of the domain using formal ontology representations such as OWL/RDF. A large-scale representation of abstract concepts such as actions, time, physical objects and beliefs would be an example of ontological engineering. Ontology engineering is one of the areas of applied ontology, and can be seen as an application of philosophical ontology. Core ideas and objectives of ontology engineering are also central in conceptual modeling.
TriX is a serialization format for RDF graphs. It is an XML format for serializing Named Graphs and RDF Datasets which offers a compact and readable alternative to the XML-based RDF/XML syntax. It was jointly created by HP Labs and Nokia.
XHTML+RDFa is an extended version of the XHTML markup language for supporting RDF through a collection of attributes and processing rules in the form of well-formed XML documents. XHTML+RDFa is one of the techniques used to develop Semantic Web content by embedding rich semantic markup. Version 1.1 of the language is a superset of XHTML 1.1, integrating the attributes according to RDFa Core 1.1. In other words, it is an RDFa support through XHTML Modularization.
Knowledge extraction is the creation of knowledge from structured and unstructured sources. The resulting knowledge needs to be in a machine-readable and machine-interpretable format and must represent knowledge in a manner that facilitates inferencing. Although it is methodically similar to information extraction (NLP) and ETL, the main criterion is that the extraction result goes beyond the creation of structured information or the transformation into a relational schema. It requires either the reuse of existing formal knowledge or the generation of a schema based on the source data.
JSON-LD is a method of encoding linked data using JSON. One goal for JSON-LD was to require as little effort as possible from developers to transform their existing JSON to JSON-LD. JSON-LD allows data to be serialized in a way that is similar to traditional JSON. It was initially developed by the JSON for Linking Data Community Group before being transferred to the RDF Working Group for review, improvement, and standardization, and is currently maintained by the JSON-LD Working Group. JSON-LD is a World Wide Web Consortium Recommendation.