Related Research Articles

Meiosis (; from Ancient Greek μείωσις 'lessening', is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, the sperm or egg cells. It involves two rounds of division that ultimately result in four cells, each with only one copy of each chromosome. Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and a female will fuse to create a zygote, a cell with two copies of each chromosome again.

Mitosis is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis is an equational division which gives rise to genetically identical cells in which the total number of chromosomes is maintained. Mitosis is preceded by the S phase of interphase and is followed by telophase and cytokinesis, which divide the cytoplasm, organelles, and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis altogether define the mitotic phase of a cell cycle—the division of the mother cell into two daughter cells genetically identical to each other.

Cell division is the process by which a parent cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukaryotes, there are two distinct types of cell division: a vegetative division (mitosis), producing daughter cells genetically identical to the parent cell, and a cell division that produces haploid gametes for sexual reproduction (meiosis), reducing the number of chromosomes from two of each type in the diploid parent cell to one of each type in the daughter cells. Mitosis is a part of the cell cycle, in which, replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. In general, mitosis is preceded by the S stage of interphase and is followed by telophase and cytokinesis; which divides the cytoplasm, organelles, and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis all together define the M phase of an animal cell cycle—the division of the mother cell into two genetically identical daughter cells. To ensure proper progression through the cell cycle, DNA damage is detected and repaired at various checkpoints throughout the cycle. These checkpoints can halt progression through the cell cycle by inhibiting certain cyclin-CDK complexes. Meiosis undergoes two divisions resulting in four haploid daughter cells. Homologous chromosomes are separated in the first division of meiosis, such that each daughter cell has one copy of each chromosome. These chromosomes have already been replicated and have two sister chromatids which are then separated during the second division of meiosis. Both of these cell division cycles are used in the process of sexual reproduction at some point in their life cycle. Both are believed to be present in the last eukaryotic common ancestor.
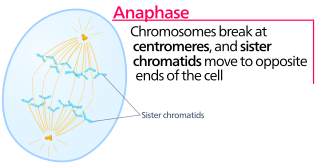
Anaphase is the stage of mitosis after the process of metaphase, when replicated chromosomes are split and the newly-copied chromosomes are moved to opposite poles of the cell. Chromosomes also reach their overall maximum condensation in late anaphase, to help chromosome segregation and the re-formation of the nucleus.

In cell biology, the spindle apparatus is the cytoskeletal structure of eukaryotic cells that forms during cell division to separate sister chromatids between daughter cells. It is referred to as the mitotic spindle during mitosis, a process that produces genetically identical daughter cells, or the meiotic spindle during meiosis, a process that produces gametes with half the number of chromosomes of the parent cell.
Cyclins are proteins that control the progression of a cell through the cell cycle by activating cyclin-dependent kinases (CDK).

The spindle checkpoint, also known as the metaphase-to-anaphase transition, the spindle assembly checkpoint (SAC), the metaphase checkpoint, or the mitotic checkpoint, is a cell cycle checkpoint during metaphase of mitosis or meiosis that prevents the separation of the duplicated chromosomes (anaphase) until each chromosome is properly attached to the spindle. To achieve proper segregation, the two kinetochores on the sister chromatids must be attached to opposite spindle poles. Only this pattern of attachment will ensure that each daughter cell receives one copy of the chromosome. The defining biochemical feature of this checkpoint is the stimulation of the anaphase-promoting complex by M-phase cyclin-CDK complexes, which in turn causes the proteolytic destruction of cyclins and proteins that hold the sister chromatids together.

A kinetochore is a disc-shaped protein structure associated with duplicated chromatids in eukaryotic cells where the spindle fibers attach during cell division to pull sister chromatids apart. The kinetochore assembles on the centromere and links the chromosome to microtubule polymers from the mitotic spindle during mitosis and meiosis. The term kinetochore was first used in a footnote in a 1934 Cytology book by Lester W. Sharp and commonly accepted in 1936. Sharp's footnote reads: "The convenient term kinetochore has been suggested to the author by J. A. Moore", likely referring to John Alexander Moore who had joined Columbia University as a freshman in 1932.
Monopolin is a protein complex that in budding yeast is composed of the four proteins CSM1, HRR25, LRS4, and MAM1. Monopolin is required for the segregation of homologous centromeres to opposite poles of a dividing cell during anaphase I of meiosis. This occurs by bridging DSN1 kinetochore proteins to sister kinetochores within the centromere to physically fuse them and allow for the microtubules to pull each homolog toward opposite mitotic spindles.

Cohesin is a protein complex that mediates sister chromatid cohesion, homologous recombination, and DNA looping. Cohesin is formed of SMC3, SMC1, SCC1 and SCC3. Cohesin holds sister chromatids together after DNA replication until anaphase when removal of cohesin leads to separation of sister chromatids. The complex forms a ring-like structure and it is believed that sister chromatids are held together by entrapment inside the cohesin ring. Cohesin is a member of the SMC family of protein complexes which includes Condensin, MukBEF and SMC-ScpAB.
Mad2 is an essential spindle checkpoint protein. The spindle checkpoint system is a regulatory system that restrains progression through the metaphase-to-anaphase transition. The Mad2 gene was first identified in the yeast S. cerevisiae in a screen for genes which when mutated would confer sensitivity to microtubule poisons. The human orthologues of Mad2 were first cloned in a search for human cDNAs that would rescue the microtubule poison-sensitivity of a yeast strain in which a kinetochore binding protein was missing. The protein was shown to be present at unattached kinetochores and antibody inhibition studies demonstrated it was essential to execute a block in the metaphase-to-anaphase transition in response to the microtubule poison nocodazole. Subsequent cloning of the Xenopus laevis orthologue, facilitated by the sharing of the human sequence, allowed for the characterization of the mitotic checkpoint in egg extracts.

Aurora kinase B is a protein that functions in the attachment of the mitotic spindle to the centromere and in cytokinesis.
Anaphase lag is a consequence of an event during cell division where sister chromatids do not properly separate from each other because of improper spindle formation. The chromosome or chromatid does not properly migrate during anaphase and the daughter cells will lose some genetic information. It is one of many causes of aneuploidy. This event can occur during both meiosis and mitosis with unique repercussions. In either case, anaphase lag will cause one daughter cell to receive a complete set of chromosomes while the other lacks one paired set of chromosomes, creating a form of monosomy. Whether the cell survives depends on which sister chromatid was lost and the background genomic state of the cell. The passage of abnormal numbers of chromosomes will have unique consequences with regards to mosaicism and development as well as the progression and heterogeneity of cancers.
Polo-like kinases (Plks) are regulatory serine/threonine kinases of the cell cycle involved in mitotic entry, mitotic exit, spindle formation, cytokinesis, and meiosis. Only one Plk is found in the genomes of the fly Drosophila melanogaster (Polo), budding yeast (Cdc5) and fission yeast (Plo1). Vertebrates and other animals, however, have many Plk family members including Plk1, Plk2/Snk, Plk3/Prk/FnK, Plk4/Sak and Plk5. Of the vertebrate Plk family members, the mammalian Plk1 has been most extensively studied. During mitosis and cytokinesis, Plks associate with several structures including the centrosome, kinetochores, and the central spindle.

Mitotic checkpoint serine/threonine-protein kinase BUB1 also known as BUB1 is an enzyme that in humans is encoded by the BUB1 gene.

Mitotic checkpoint protein BUB3 is a protein that in humans is encoded by the BUB3 gene.
Syntelic attachment occurs when both sister chromosomes are attached to a single spindle pole.

Mad1 is a non-essential protein which in yeast has a function in the spindle assembly checkpoint (SAC). This checkpoint monitors chromosome attachment to spindle microtubules and prevents cells from starting anaphase until the spindle is built up. The name Mad refers to the observation that mutant cells are mitotic arrest deficient (MAD) during microtubule depolymerization. Mad1 recruits the anaphase inhibitor Mad2 to unattached kinetochores and is essential for Mad2-Cdc20 complex formation in vivo but not in vitro. In vivo, Mad1 acts as a competitive inhibitor of the Mad2-Cdc20 complex. Mad1 is phosphorylated by Mps1 which then leads together with other activities to the formation of the mitotic checkpoint complex (MCC). Thereby it inhibits the activity of the anaphase-promoting complex/cyclosome (APC/C). Homologues of Mad1 are conserved in eukaryotes from yeast to mammals.
In molecular biology, the protein domain named the Shugoshin N-terminal coiled-coil region is a domain found on the N-terminal region of the Shugoshin protein in eukaryotes. It has a role in attaching to the kinetochores, structures on the chromatids where microtubules attach. Shugoshin has a conserved coiled-coil N-terminal domain and a highly conserved C-terminal region. Shugoshin is a crucial target of Bub1 kinase that plays a central role in the cohesion of chromosomes during cell division.
The XMAP215/Dis1 family is a highly conserved group of microtubule-associated proteins (MAPs) in eukaryotic organisms. These proteins are unique MAPs because they primarily interact with the growing-end (plus-end) of microtubules. This special property classifies this protein family as plus-end tracking proteins (+TIPs).
References
- ↑ C.B. O'Connell; A. Khodjakov & B.F. McEwen (2012). "Kinetochore flexibility: creating a dynamic chromosome-spindle interface". Current Opinion in Cell Biology. 24 (1): 40–47. doi:10.1016/j.ceb.2011.12.008. PMC 3507511 . PMID 22221609.
- ↑ G. Gay; T. Courtheoux; C. reyes; S. Tournier & B.F. McEwen (2012). "A stochastic model of kinetochore-microtubule attachment accurately describes fission yeast chromosome segregation". Journal of Cell Biology. 196 (6): 757–774. doi:10.1083/jcb.201107124. PMC 3308688 . PMID 22412019.
- 1 2 3 T.S. Kitajima; M. Ohsugi & J. Ellenberg (2011). "Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes". Cell. 146 (4): 568–581. doi: 10.1016/j.cell.2011.07.031 . PMID 21854982.
- 1 2 J.R. Geert; L. Kops; A.T. Saurin & P. Meraldi (2010). "Finding the middle ground: how kinetochores power chromosome congression". Cellular and Molecular Life Sciences. 67 (13): 2145–2161. doi:10.1007/s00018-010-0321-y. PMC 2883098 . PMID 20232224.