Related Research Articles
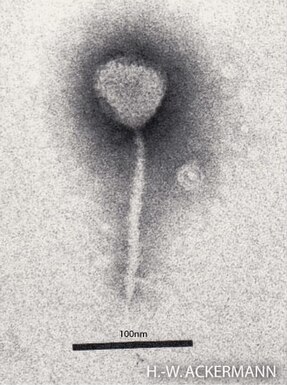
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

Shiga toxins are a family of related toxins with two major groups, Stx1 and Stx2, expressed by genes considered to be part of the genome of lambdoid prophages. The toxins are named after Kiyoshi Shiga, who first described the bacterial origin of dysentery caused by Shigella dysenteriae. Shiga-like toxin (SLT) is a historical term for similar or identical toxins produced by Escherichia coli. The most common sources for Shiga toxin are the bacteria S. dysenteriae and some serotypes of Escherichia coli (STEC), which includes serotypes O157:H7, and O104:H4.

The lytic cycle is one of the two cycles of viral reproduction, the other being the lysogenic cycle. The lytic cycle results in the destruction of the infected cell and its membrane. Bacteriophages that only use the lytic cycle are called virulent phages.

Tectiviridae is a family of viruses with seven species in three genera. Bacteria serve as natural hosts. Tectiviruses have no head-tail structure, but are capable of producing tail-like tubes of ~ 60×10 nm upon adsorption or after chloroform treatment. The name is derived from Latin tectus.
The hok/sok system is a postsegregational killing mechanism employed by the R1 plasmid in Escherichia coli. It was the first type I toxin-antitoxin pair to be identified through characterisation of a plasmid-stabilising locus. It is a type I system because the toxin is neutralised by a complementary RNA, rather than a partnered protein.
Virulence-related outer membrane proteins are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.
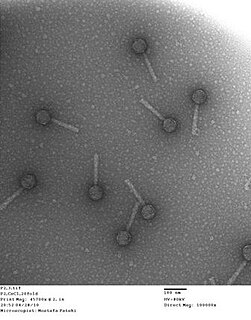
Bacteriophage P2, scientific name Escherichia virus P2, is a temperate phage that infects E. coli. It is a tailed virus with a contractile sheath and is thus classified in the genus Peduovirus, subfamily Peduovirinae, family Myoviridae within order Caudovirales. This genus of viruses includes many P2-like phages as well as the satellite phage P4.
Holins are a diverse group of small proteins produced by dsDNA bacteriophages in order to trigger and control the degradation of the host's cell wall at the end of the lytic cycle. Holins form pores in the host's cell membrane, allowing lysins to reach and degrade peptidoglycan, a component of bacterial cell walls. Holins have been shown to regulate the timing of lysis with great precision. Over 50 unrelated gene families encode holins, making them the most diverse group of proteins with common function. Together with lysins, holins are being studied for their potential use as antibacterial agents.
Shigatoxigenic Escherichia coli (STEC) and verotoxigenic E. coli (VTEC) are strains of the bacterium Escherichia coli that produce either Shiga toxin or Shiga-like toxin (verotoxin). Only a minority of the strains cause illness in humans. The ones that do are collectively known as enterohemorrhagic E. coli (EHEC) and are major causes of foodborne illness. When infecting humans, they often cause gastroenteritis, enterocolitis, and bloody diarrhea and sometimes cause a severe complication called hemolytic-uremic syndrome (HUS). The group and its subgroups are known by various names. They are distinguished from other strains of intestinal pathogenic E. coli including enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), and diffusely adherent E. coli (DAEC).

cII or transcriptional activator II is a DNA-binding protein and important transcription factor in the life cycle of lambda phage. It is encoded in the lambda phage genome by the 291 base pair cII gene. cII plays a key role in determining whether the bacteriophage will incorporate its genome into its host and lie dormant (lysogeny), or replicate and kill the host (lysis).
The Phage 21 S Family is a member of the Holin Superfamily II.
The T7 Holin family is a member of the Holin Superfamily II. Members of this family are predominantly found in Caudovirales and Proteobacteria. They typically have only 1 transmembrane segment (TMS) and vary from 60 to 130 amino acyl residues in length. A representative list of proteins belonging to this family can be found in the Transporter Classification Database.
The HP1 Holin Family is a member of the Holin Superfamily II. Proteins in this family are typically found to contain two transmembrane segments (TMSs) and range between 70 and 80 amino acyl residues (aas) in length. A representative list of proteins belonging to the HP1 holin family can be found in the Transporter Classification Database.
The Pseudomonas phage F116 holin is a non-characterized holin homologous to one in Neisseria gonorrheae that has been characterized. This protein is the prototype of the Pseudomonasphage F116 holin family, which is a member of the Holin Superfamily II. Bioinformatic analysis of the genome sequence of N. gonorrhoeae revealed the presence of nine probable prophage islands. The genomic sequence of FA1090 identified five genomic regions that are related to dsDNA lysogenic phage. The DNA sequences from NgoPhi1, NgoPhi2 and NgoPhi3 contained regions of identity. A region of NgoPhi2 showed high similarity with the Pseudomonas aeruginosa generalized transducing phage F116. NgoPhi1 and NgoPhi2 encode functionally active phages. The holin gene of NgoPhi1, when expressed in E. coli, could substitute for the phage lambda S gene.
The Lambda Holin S Family is a group of integral membrane transporter proteins belonging to the Holin Superfamily III. Members of this family generally consist of the characteristic three transmembrane segments (TMSs) and are of 110 amino acyl residues (aas) in length, on average. A representative list of members belonging to this family can be found in the Transporter Classification Database.
The PRD1 Phage P35 Holin Family is a member of Holin Superfamily III. The prototype for this family is the lipid-containing PRD1 enterobacterial phage holin protein P35 encoded by gene XXXV (orfT). It is a component of a typical holin-endolysin system which functions to lyse the host bacterial cell.
The Holin Hol44 (Hol44) Family is a group of transporters belonging to the Holin Superfamily V. A representative list of proteins belonging to the Hol44 family from caudovirales and firmicutes can be found in the Transporter Classification Database.
The T4 Holin Family is a group of putative pore-forming proteins that does not belong to one of the seven holin superfamilies. T-even phage such as T4 use a holin-endolysin system for host cell lysis. Although the endolysin of phage T4 encoded by the e gene was identified in 1961, the holin was not characterized until 2001. A representative list of proteins belonging to the T4 holin family can be found in the Transporter Classification Database.
The Lactococcus lactis Phage r1t Holin Family is a family of putative pore-forming proteins that typically range in size between about 65 and 95 amino acyl residues (aas) in length, although a few r1t holins have been found to be significantly larger. Phage r1t holins exhibit between 2 and 4 transmembrane segments (TMSs), with the 4 TMS proteins resulting from an intragenic duplication of a 2 TMS region. A representative list of the proteins belonging to the r1t holin family can be found in the Transporter Classification Database.
The Actinobacterial Phage Holin (APH) Family is a fairly large family of proteins between 105 and 180 amino acyl residues in length, typically exhibiting a single transmembrane segment (TMS) near the N-terminus. A representative list of proteins belonging to the APH family can be found in the Transporter Classification Database.
References
- ↑ "1.E.17 The BlyA Holin (BlyA Holin) Family". Transporter Classification Database. Retrieved 2016-03-28.
- 1 2 Damman, C. J.; Eggers, C. H.; Samuels, D. S.; Oliver, D. B. (2000-12-01). "Characterization of Borrelia burgdorferi BlyA and BlyB proteins: a prophage-encoded holin-like system". Journal of Bacteriology. 182 (23): 6791–6797. doi:10.1128/jb.182.23.6791-6797.2000. ISSN 0021-9193. PMC 111423 . PMID 11073925.
- ↑ Ludwig, Albrecht; von Rhein, Christine; Mischke, Annette; Brade, Volker (2008-07-01). "Release of latent ClyA cytolysin from Escherichia coli mediated by a bacteriophage-associated putative holin (BlyA) from Borrelia burgdorferi". International Journal of Medical Microbiology. 298 (5–6): 473–481. doi:10.1016/j.ijmm.2007.07.014. ISSN 1618-0607. PMID 17897882.
As of this edit, this article uses content from "1.E.17 The BlyA Holin (BlyA Holin) Family" , which is licensed in a way that permits reuse under the Creative Commons Attribution-ShareAlike 3.0 Unported License, but not under the GFDL. All relevant terms must be followed.