
Metabolomics is the scientific study of chemical processes involving metabolites, the small molecule substrates, intermediates and products of cell metabolism. Specifically, metabolomics is the "systematic study of the unique chemical fingerprints that specific cellular processes leave behind", the study of their small-molecule metabolite profiles. The metabolome represents the complete set of metabolites in a biological cell, tissue, organ or organism, which are the end products of cellular processes. Messenger RNA (mRNA), gene expression data and proteomic analyses reveal the set of gene products being produced in the cell, data that represents one aspect of cellular function. Conversely, metabolic profiling can give an instantaneous snapshot of the physiology of that cell, and thus, metabolomics provides a direct "functional readout of the physiological state" of an organism. There are indeed quantifiable correlations between the metabolome and the other cellular ensembles, which can be used to predict metabolite abundances in biological samples from, for example mRNA abundances. One of the ultimate challenges of systems biology is to integrate metabolomics with all other -omics information to provide a better understanding of cellular biology.

The metabolome refers to the complete set of small-molecule chemicals found within a biological sample. The biological sample can be a cell, a cellular organelle, an organ, a tissue, a tissue extract, a biofluid or an entire organism. The small molecule chemicals found in a given metabolome may include both endogenous metabolites that are naturally produced by an organism as well as exogenous chemicals that are not naturally produced by an organism.

KEGG is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. KEGG is utilized for bioinformatics research and education, including data analysis in genomics, metagenomics, metabolomics and other omics studies, modeling and simulation in systems biology, and translational research in drug development.
Fluxomics describes the various approaches that seek to determine the rates of metabolic reactions within a biological entity. While metabolomics can provide instantaneous information on the metabolites in a biological sample, metabolism is a dynamic process. The significance of fluxomics is that metabolic fluxes determine the cellular phenotype. It has the added advantage of being based on the metabolome which has fewer components than the genome or proteome.

The Human Metabolome Database (HMDB) is a comprehensive, high-quality, freely accessible, online database of small molecule metabolites found in the human body. Created by the Human Metabolome Project funded by Genome Canada. One of the first dedicated metabolomics databases, the HMDB facilitates human metabolomics research, including the identification and characterization of human metabolites using NMR spectroscopy, GC-MS spectrometry and LC/MS spectrometry. To aid in this discovery process, the HMDB contains three kinds of data: 1) chemical data, 2) clinical data, and 3) molecular biology/biochemistry data. The chemical data includes 41,514 metabolite structures with detailed descriptions along with nearly 10,000 NMR, GC-MS and LC/MS spectra.
The Small Molecule Pathway Database (SMPDB) is a comprehensive, high-quality, freely accessible, online database containing more than 600 small molecule pathways found in humans. SMPDB is designed specifically to support pathway elucidation and pathway discovery in metabolomics, transcriptomics, proteomics and systems biology. It is able to do so, in part, by providing colorful, detailed, fully searchable, hyperlinked diagrams of five types of small molecule pathways: 1) general human metabolic pathways; 2) human metabolic disease pathways; 3) human metabolite signaling pathways; 4) drug-action pathways and 5) drug metabolism pathways. SMPDB pathways may be navigated, viewed and zoomed interactively using a Google Maps-like interface. All SMPDB pathways include information on the relevant organs, subcellular compartments, protein cofactors, protein locations, metabolite locations, chemical structures and protein quaternary structures. Each small molecule in SMPDB is hyperlinked to detailed descriptions contained in the HMDB or DrugBank and each protein or enzyme complex is hyperlinked to UniProt. Additionally, all SMPDB pathways are accompanied with detailed descriptions and references, providing an overview of the pathway, condition or processes depicted in each diagram. Users can browse the SMPDB or search its contents by text searching, sequence searching, or chemical structure searching. More powerful queries are also possible including searching with lists of gene or protein names, drug names, metabolite names, GenBank IDs, Swiss-Prot IDs, Agilent or Affymetrix microarray IDs. These queries will produce lists of matching pathways and highlight the matching molecules on each of the pathway diagrams. Gene, metabolite and protein concentration data can also be visualized through SMPDB's mapping interface.

The Golm Metabolome Database (GMD) is a gas chromatography (GC) – mass spectrometry (MS) reference library dedicated to metabolite profiling experiments and comprises mass spectral and retention index (RI) information for non-annotated mass spectral tags together with data of a multitude of already identified metabolites and reference substances. The GMD is hosted at the Max Planck Institute of Molecular Plant Physiology in Golm district of Potsdam, Germany.
The Yeast Metabolome Database (YMDB) is a comprehensive, high-quality, freely accessible, online database of small molecule metabolites found in or produced by Saccharomyces cerevisiae. The YMDB was designed to facilitate yeast metabolomics research, specifically in the areas of general fermentation as well as wine, beer and fermented food analysis. YMDB supports the identification and characterization of yeast metabolites using NMR spectroscopy, GC-MS spectrometry and Liquid chromatography–mass spectrometry. The YMDB contains two kinds of data: 1) chemical data and 2) molecular biology/biochemistry data. The chemical data includes 2027 metabolite structures with detailed metabolite descriptions along with nearly 4000 NMR, GC-MS and LC/MS spectra.
Metabolite Set Enrichment Analysis (MSEA) is a method designed to help metabolomics researchers identify and interpret patterns of metabolite concentration changes in a biologically meaningful way. It is conceptually similar to another widely used tool developed for transcriptomics called Gene Set Enrichment Analysis or GSEA. GSEA uses a collection of predefined gene sets to rank the lists of genes obtained from gene chip studies. By using this “prior knowledge” about gene sets researchers are able to readily identify significant and coordinated changes in gene expression data while at the same time gaining some biological context. MSEA does the same thing by using a collection of predefined metabolite pathways and disease states obtained from the Human Metabolome Database. MSEA is offered as a service both through a stand-alone web server and as part of a larger metabolomics analysis suite called MetaboAnalyst.
The E. coli Metabolome Database (ECMDB) is a comprehensive, high-quality, freely accessible, online database of small molecule metabolites found in or produced by Escherichia coli. Escherichia coli is perhaps the best studied bacterium on earth and has served as the "model microbe" in microbiology research for more than 60 years. The ECMDB is essentially an E. coli "omics" encyclopedia containing detailed data on E. coli's genome, proteome and its metabolome. ECMDB is part of a suite of organism-specific metabolomics databases that includes DrugBank, HMDB, YMDB and SMPDB. As a metabolomics resource, the ECMDB is designed to facilitate research in the area gut/microbiome metabolomics and environmental metabolomics. The ECMDB contains two kinds of data: 1) chemical data and 2) molecular biology and/or biochemical data. The chemical data includes more than 2700 metabolite structures with detailed metabolite descriptions along with nearly 5000 NMR, GC-MS and LC-MS spectra corresponding to these metabolites. The biochemical data includes nearly 1600 protein sequences and more than 3100 biochemical reactions that are linked to these metabolite entries. Each metabolite entry in the ECMDB contains more than 80 data fields with approximately 65% of the information being devoted to chemical data and the other 35% of the information devoted to enzymatic or biochemical data. Many data fields are hyperlinked to other databases. The ECMDB also has a variety of structure and pathway viewing applets. The ECMDB database offers a number of text, sequence, spectral, chemical structure and relational query searches. These are described in more detail below.

Estriol glucuronide (E3G), or oestriol glucuronide, also known as estriol monoglucuronide, as well as estriol 16α-β-D-glucosiduronic acid, is a natural, steroidal estrogen and the glucuronic acid conjugate of estriol. It occurs in high concentrations in the urine of pregnant women as a reversibly formed metabolite of estriol. Estriol glucuronide is a prodrug of estriol, and was the major component of Progynon and Emmenin, estrogenic products manufactured from the urine of pregnant women that were introduced in the 1920s and 1930s and were the first orally active estrogens. Emmenin was succeeded by Premarin, which is sourced from the urine of pregnant mares and was introduced in 1941. Premarin replaced Emmenin due to the fact that it was easier and less expensive to produce.

Androsterone glucuronide (ADT-G) is a major circulating and urinary metabolite of testosterone and dihydrotestosterone (DHT). It accounts for 93% of total androgen glucuronides in women. ADT-G is formed from androsterone by UDP-glucuronosyltransferases, with the major enzymes being UGT2B15 and UGT2B17. It is a marker of acne in women while androstanediol glucuronide is a marker of hirsutism in women.
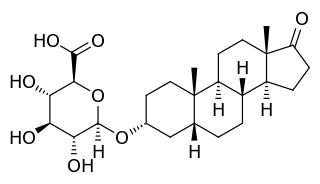
Etiocholanolone glucuronide (ETIO-G) is an endogenous, naturally occurring metabolite of testosterone. It is formed in the liver from etiocholanolone by UDP-glucuronyltransferases. ETIO-G has much higher water solubility than etiocholanolone and is eventually excreted in the urine via the kidneys. Along with androsterone glucuronide, it is one of the major inactive metabolites of testosterone.

4-Hydroxyestradiol (4-OHE2), also known as estra-1,3,5(10)-triene-3,4,17β-triol, is an endogenous, naturally occurring catechol estrogen and a minor metabolite of estradiol. It is estrogenic, similarly to many other hydroxylated estrogen metabolites such as 2-hydroxyestradiol, 16α-hydroxyestrone, estriol (16α-hydroxyestradiol), and 4-hydroxyestrone but unlike 2-hydroxyestrone.
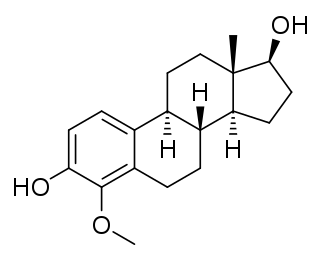
4-Methoxyestradiol (4-ME2) is an endogenous, naturally occurring methoxylated catechol estrogen and metabolite of estradiol that is formed by catechol O-methyltransferase via the intermediate 4-hydroxyestradiol. It has estrogenic activity similarly to estrone and 4-hydroxyestrone.

Androsterone sulfate, also known as 3α-hydroxy-5α-androstan-17-one 3α-sulfate, is an endogenous, naturally occurring steroid and one of the major urinary metabolites of androgens. It is a steroid sulfate which is formed from sulfation of androsterone by the steroid sulfotransferase SULT2A1 and can be desulfated back into androsterone by steroid sulfatase.
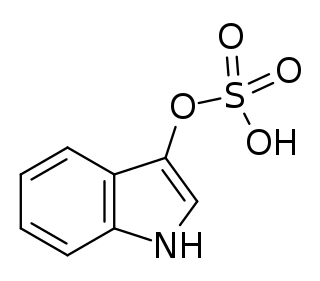
Indoxyl sulfate, also known as 3-indoxylsulfate and 3-indoxylsulfuric acid, is a metabolite of dietary L-tryptophan that acts as a cardiotoxin and uremic toxin. High concentrations of indoxyl sulfate in blood plasma are known to be associated with the development and progression of chronic kidney disease and vascular disease in humans. As a uremic toxin, it stimulates glomerular sclerosis and renal interstitial fibrosis.

Etiocholanedione, also known as 5β-androstanedione or as etiocholane-3,17-dione, is a naturally occurring etiocholane (5β-androstane) steroid and an endogenous metabolite of androgens like testosterone, dihydrotestosterone, dehydroepiandrosterone (DHEA), and androstenedione. It is the C5 epimer of androstanedione (5α-androstanedione). Although devoid of androgenic activity like other 5β-reduced steroids, etiocholanedione has some biological activity of its own. The compound has been found to possess potent haematopoietic effects in a variety of models. In addition, it has been found to promote weight loss in animals and in a double-blind, placebo-controlled clinical study in humans conducted in 1993. These effects are said to be similar to those of DHEA. Unlike DHEA however, etiocholanedione cannot be metabolized further into steroid hormones like androgens and estrogens.

David S. Wishart FRSC is a Canadian researcher and a Distinguished University Professor in the Department of Biological Sciences and the Department of Computing Science at the University of Alberta, where he has been since 1995. Wishart also holds cross appointments in the Faculty of Pharmacy and Pharmaceutical Sciences and the Department of Laboratory Medicine and Pathology in the Faculty of Medicine and Dentistry. Additionally, Wishart holds a joint appointment in metabolomics at the Pacific Northwest National Laboratory in Richland, Washington. Wishart is well known for his pioneering contributions to the fields of protein NMR spectroscopy, bioinformatics, cheminformatics and metabolomics. In 2011, Wishart founded and currently serves as a co-director of he Metabolomics Innovation Centre (TMIC), which is Canada's national metabolomics laboratory.














