Hox genes, a subset of homeobox genes, are a group of related genes that specify regions of the body plan of an embryo along the head-tail axis of animals. Hox proteins encode and specify the characteristics of 'position', ensuring that the correct structures form in the correct places of the body. For example, Hox genes in insects specify which appendages form on a segment, and Hox genes in vertebrates specify the types and shape of vertebrae that will form. In segmented animals, Hox proteins thus confer segmental or positional identity, but do not form the actual segments themselves.

Ultrabithorax (Ubx) is a homeobox gene found in insects, and is used in the regulation of patterning in morphogenesis. There are many possible products of this gene, which function as transcription factors. Ubx is used in the specification of serially homologous structures, and is used at many levels of developmental hierarchies. In Drosophila melanogaster it is expressed in the third thoracic (T3) and first abdominal (A1) segments and represses wing formation. The Ubx gene regulates the decisions regarding the number of wings and legs the adult flies will have. The developmental role of the Ubx gene is determined by the splicing of its product, which takes place after translation of the gene. The specific splice factors of a particular cell allow the specific regulation of the developmental fate of that cell, by making different splice variants of transcription factors. In D. melanogaster, at least six different isoforms of Ubx exist.
Polycomb-group proteins are a family of protein complexes first discovered in fruit flies that can remodel chromatin such that epigenetic silencing of genes takes place. Polycomb-group proteins are well known for silencing Hox genes through modulation of chromatin structure during embryonic development in fruit flies. They derive their name from the fact that the first sign of a decrease in PcG function is often a homeotic transformation of posterior legs towards anterior legs, which have a characteristic comb-like set of bristles.

In molecular biology, U3 snoRNA is a non-coding RNA found predominantly in the nucleolus. U3 has C/D box motifs that technically make it a member of the box C/D class of snoRNAs; however, unlike other C/D box snoRNAs, it has not been shown to direct 2'-O-methylation of other RNAs. Rather, U3 is thought to guide site-specific cleavage of ribosomal RNA (rRNA) during pre-rRNA processing.
In molecular biology, Small nucleolar RNA R20 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA R20 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA R20 was identified in a screen of Arabidopsis thaliana.

In molecular biology, Small nucleolar RNA R28 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA R28 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA R28 was identified in a screen of Arabidopsis thaliana.

In molecular biology, U8 small nucleolar RNA is the RNA component of a small RNA:protein complex which is required for biogenesis of mature large subunit ribosomal RNAs, 5.8S and 28S rRNAs.

Polycomb complex protein BMI-1 also known as polycomb group RING finger protein 4 (PCGF4) or RING finger protein 51 (RNF51) is a protein that in humans is encoded by the BMI1 gene. BMI1 is a polycomb ring finger oncogene.

Histone-lysine N-methyltransferase SUV39H1 is an enzyme that in humans is encoded by the SUV39H1 gene.

Histone acetyltransferase KAT2A is an enzyme that in humans is encoded by the KAT2A gene.

E3 ubiquitin-protein ligase RING2 is an enzyme that in humans is encoded by the RNF2 gene.

Histone RNA hairpin-binding protein or stem-loop binding protein (SLBP) is a protein that in humans is encoded by the SLBP gene.

Polycomb protein EED is a protein that in humans is encoded by the EED gene.

WD repeat-containing protein 5 is a protein that in humans is encoded by the WDR5 gene.

Set1/Ash2 histone methyltransferase complex subunit ASH2 is an enzyme that in humans is encoded by the ASH2L gene.

ASH1L is a histone-lysine N-methyltransferase enzyme encoded by the ASH1L gene located at chromosomal band 1q22. ASH1L is the human homolog of Drosophila Ash1.
Trithorax-group proteins (TrxG) are a heterogeneous collection of proteins whose main action is to maintain gene expression. They can be categorized into three general classes based on molecular function:
- histone-modifying TrxG proteins
- chromatin-remodeling TrxG proteins
- DNA-binding TrxG proteins,
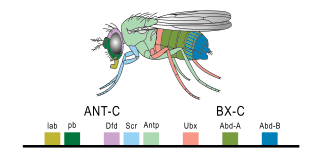
The Bithorax complex (BX-C) is one of two Drosophila melanogaster homeotic gene complexes, located on the right arm of chromosome 3. It is responsible for the differentiation of the posterior two-thirds of the fly by the regulation of three genes within the complex: Ultrabithorax (Ubx), abdominal A (abd-A), and Abdominal B (Abd-B).
Homeotic selector genes confer segment identity in Drosophila. They encode homeodomain proteins which interact with Hox and other homeotic genes to initiate segment-specific gene regulation. Homeodomain proteins are transcription factors that share a DNA-binding domain called the homeodomain. Changes in the expression and function of homeotic genes are responsible for the changes in the morphology of the limbs of arthropods as well as in the axial skeletons of vertebrates. Mutations in homeotic selector genes do not lead to elimination of a segment or pattern, but instead cause the segment to develop incorrectly.

In molecular biology the MYND-type zinc finger domain is a conserved protein domain. The MYND domain is present in a large group of proteins that includes RP-8 (PDCD2), Nervy, and predicted proteins from Drosophila, mammals, Caenorhabditis elegans, yeast, and plants. The MYND domain consists of a cluster of cysteine and histidine residues, arranged with an invariant spacing to form a potential zinc-binding motif. Mutating conserved cysteine residues in the DEAF-1 MYND domain does not abolish DNA binding, which suggests that the MYND domain might be involved in protein-protein interactions. Indeed, the MYND domain of ETO/MTG8 interacts directly with the N-CoR and SMRT co-repressors. Aberrant recruitment of co-repressor complexes and inappropriate transcriptional repression is believed to be a general mechanism of leukemogenesis caused by the t(8;21) translocations that fuse ETO with the acute myelogenous leukemia 1 (AML1) protein. ETO has been shown to be a co-repressor recruited by the promyelocytic leukemia zinc finger (PLZF) protein. A divergent MYND domain present in the adenovirus E1A binding protein BS69 was also shown to interact with N-CoR and mediate transcriptional repression. The current evidence suggests that the MYND motif in mammalian proteins constitutes a protein-protein interaction domain that functions as a co-repressor-recruiting interface.
















