
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences.
Native Chemical Ligation (NCL) is an important extension of the chemical ligation concept for constructing a larger polypeptide chain by the covalent condensation of two or more unprotected peptides segments. Native chemical ligation is the most effective method for synthesizing native or modified proteins of typical size.
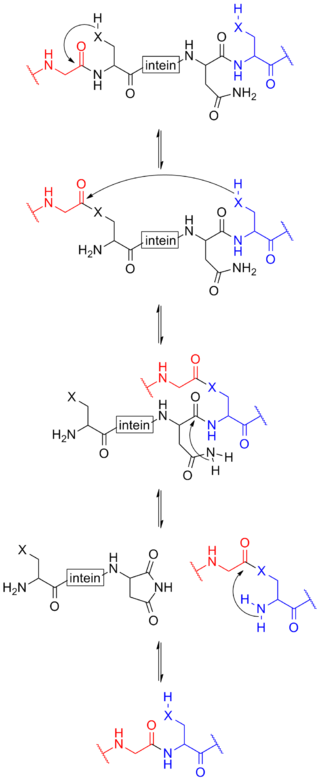
Protein splicing is an intramolecular reaction of a particular protein in which an internal protein segment is removed from a precursor protein with a ligation of C-terminal and N-terminal external proteins on both sides. The splicing junction of the precursor protein is mainly a cysteine or a serine, which are amino acids containing a nucleophilic side chain. The protein splicing reactions which are known now do not require exogenous cofactors or energy sources such as adenosine triphosphate (ATP) or guanosine triphosphate (GTP). Normally, splicing is associated only with pre-mRNA splicing. This precursor protein contains three segments—an N-extein followed by the intein followed by a C-extein. After splicing has taken place, the resulting protein contains the N-extein linked to the C-extein; this splicing product is also termed an extein.
Nonribosomal peptides (NRP) are a class of peptide secondary metabolites, usually produced by microorganisms like bacteria and fungi. Nonribosomal peptides are also found in higher organisms, such as nudibranchs, but are thought to be made by bacteria inside these organisms. While there exist a wide range of peptides that are not synthesized by ribosomes, the term nonribosomal peptide typically refers to a very specific set of these as discussed in this article.

In organic chemistry, peptide synthesis is the production of peptides, compounds where multiple amino acids are linked via amide bonds, also known as peptide bonds. Peptides are chemically synthesized by the condensation reaction of the carboxyl group of one amino acid to the amino group of another. Protecting group strategies are usually necessary to prevent undesirable side reactions with the various amino acid side chains. Chemical peptide synthesis most commonly starts at the carboxyl end of the peptide (C-terminus), and proceeds toward the amino-terminus (N-terminus). Protein biosynthesis in living organisms occurs in the opposite direction.
Cyanogen bromide is the inorganic compound with the formula (CN)Br or BrCN. It is a colorless solid that is widely used to modify biopolymers, fragment proteins and peptides, and synthesize other compounds. The compound is classified as a pseudohalogen.
Stephen B. H. Kent is a professor at the University of Chicago. While professor at the Scripps Research Institute in the early 1990s he pioneered modern ligation methods for the total chemical synthesis of proteins. He was the inventor of native chemical ligation together with his student Philip Dawson. His laboratory experimentally demonstrated the principle that chemical synthesis of a protein's polypeptide chain using mirror-image amino acids after folding results in a mirror-image protein molecule which, if an enzyme, will catalyze a chemical reaction with mirror-image stereospecificity. At the University of Chicago Kent and his junior colleagues pioneered the elucidation of protein structures by quasi-racemic & racemic crystallography.
A peptide library is a tool for studying proteins. Peptide libraries typically contain a large number of peptides that have a systematic combination of amino acids. Usually, solid phase synthesis, e.g. resin as a flat surface or beads, is used for peptide library generation. Peptide libraries are a popular tool for experiments in drug design, protein–protein interactions, and other biochemical and pharmaceutical applications.

An isopeptide bond is a type of amide bond formed between a carboxyl group of one amino acid and an amino group of another. An isopeptide bond is the linkage between the side chain amino or carboxyl group of one amino acid to the α-carboxyl, α-amino group, or the side chain of another amino acid. In a typical peptide bond, also known as eupeptide bond, the amide bond always forms between the α-carboxyl group of one amino acid and the α-amino group of the second amino acid. Isopeptide bonds are rarer than regular peptide bonds. Isopeptide bonds lead to branching in the primary sequence of a protein. Proteins formed from normal peptide bonds typically have a linear primary sequence.

mRNA display is a display technique used for in vitro protein, and/or peptide evolution to create molecules that can bind to a desired target. The process results in translated peptides or proteins that are associated with their mRNA progenitor via a puromycin linkage. The complex then binds to an immobilized target in a selection step. The mRNA-protein fusions that bind well are then reverse transcribed to cDNA and their sequence amplified via a polymerase chain reaction. The result is a nucleotide sequence that encodes a peptide with high affinity for the molecule of interest.

Pseudoproline derivatives are artificially created dipeptides to minimize aggregation during Fmoc solid-phase synthesis of peptides.
Bioconjugation is a chemical strategy to form a stable covalent link between two molecules, at least one of which is a biomolecule.

Racemic crystallography is a technique used in structural biology where crystals of a protein molecule are developed from an equimolar mixture of an L-protein molecule of natural chirality and its D-protein mirror image. L-protein molecules consist of 'left-handed' L-amino acids and the achiral amino acid glycine, whereas the mirror image D-protein molecules consist of 'right-handed' D-amino acids and glycine. Typically, both the L-protein and the D-protein are prepared by total chemical synthesis.
Glycopeptides are peptides that contain carbohydrate moieties (glycans) covalently attached to the side chains of the amino acid residues that constitute the peptide.
Protein chemical synthesis by native peptide ligation of unprotected peptide segments is an interesting complement and potential alternative to the use of living systems for producing proteins. The synthesis of proteins requires efficient native peptide ligation methods, which enable the chemoselective formation of a native peptide bond in aqueous solution between unprotected peptide segments. The most frequently used technique for synthesizing proteins is Native chemical ligation (NCL). However, alternatives are emerging, one of which is SEA Native Peptide Ligation.
Ribosomally synthesized and post-translationally modified peptides (RiPPs), also known as ribosomal natural products, are a diverse class of natural products of ribosomal origin. Consisting of more than 20 sub-classes, RiPPs are produced by a variety of organisms, including prokaryotes, eukaryotes, and archaea, and they possess a wide range of biological functions.
The α-Ketoacid-Hydroxylamine (KAHA) Amide-Forming Ligation is a chemical reaction that is used to join two unprotected fragments in peptide synthesis. It is an alternative to the Native Chemical Ligation (NCL). KAHA Ligation was developed by Jeffrey W. Bode group at ETH Zürich.
Bradley Lether Pentelute is currently a professor of chemistry at the Massachusetts Institute of Technology (MIT). His research program lies at the intersection of chemistry and biology and develops bioconjugation strategies, cytosolic delivery platforms, and rapid flow synthesis technologies to optimize the production, achieve site-specific modification, enhance stability, and modulate function of a variety of bioactive agents. His laboratory successfully modified proteins via cysteine-containing “pi-clamps” made up of a short sequence of amino acids, and delivered large biomolecules, such as various proteins and drugs, into cells via the anthrax delivery vehicle. Pentelute has also made several key contributions to automated synthesis technologies in flow. These advances includes the invention of the world's fastest polypeptide synthesizer. This system is able to form amide bonds at a more efficient rate than standard commercial equipment and has helped in the process of understanding protein folding and its mechanisms. This automated flow technology was recently used to achieve total chemical synthesis of protein chains up to 164 amino acids in length that retained the structure and function of native variants obtained by recombinant expression. The primary goal of his endeavor is to use these processes to create designer biologics that can be used to treat diseases and solve the manufacturing problem for on-demand personalized therapies, such as cancer vaccines.
The SpyTag/SpyCatcher system is a technology for irreversible conjugation of recombinant proteins. The peptide SpyTag spontaneously reacts with the protein SpyCatcher to form an intermolecular isopeptide bond between the pair. DNA sequence encoding either SpyTag or SpyCatcher can be recombinantly introduced into the DNA sequence encoding a protein of interest, forming a fusion protein. These fusion proteins can be covalently linked when mixed in a reaction through the SpyTag/SpyCatcher system.






