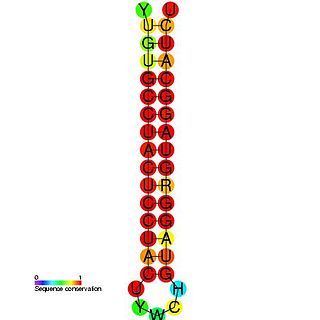
Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.
VPg is a protein that is covalently attached to the 5′ end of positive strand viral RNA and acts as a primer during RNA synthesis in a variety of virus families including Picornaviridae, Potyviridae and Caliciviridae. There are some studies showing that a possible VPg protein is also present in astroviridae, however, experimental evidence for this has not yet been provided and requires further study. The primer activity of VPg occurs when the protein becomes uridylated, providing a free hydroxyl that can be extended by the virally encoded RNA-dependent RNA polymerase. For some virus families, VPg also has a role in translation initiation by acting like a 5' mRNA cap.

Closterovirus, also known as beet yellows viral group, is a genus of viruses, in the family Closteroviridae. Plants serve as natural hosts. There are 17 species in this genus. Diseases associated with this genus include: yellowing and necrosis, particularly affecting the phloem. This genus has a probably worldwide distribution and includes among other viral species the Beet yellows virus and Citrus tristeza virus, rather economically important plant diseases. At least some species require vectors such as aphids or mealybugs for their transmission from plant to plant.
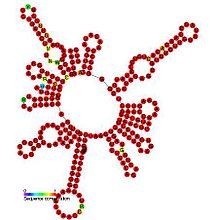
Crinivirus, formerly the lettuce infectious yellows virus group, is a genus of viruses, in the family Closteroviridae. They are linear, single-stranded positive sense RNA viruses. There are 14 species in this genus. Diseases associated with this genus include: yellowing and necrosis, particularly affecting the phloem.
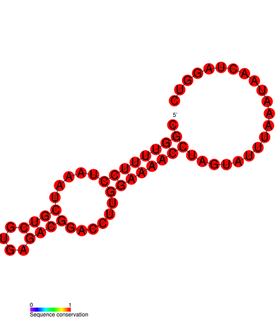
This family represents a Cardiovirus cis-acting replication element (CRE) which is located within the region encoding the capsid protein VP2 and is required for viral replication.
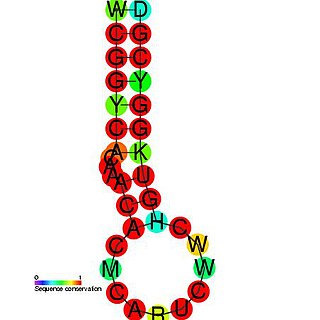
The Hepatitis C stem-loop IV is part of a putative RNA element found in the NS5B coding region. This element along with stem-loop VII, is important for colony formation, though its exact function and mechanism are unknown.

The hepatitis C virus 3′X element is an RNA element which contains three stem-loop structures that are essential for replication.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.
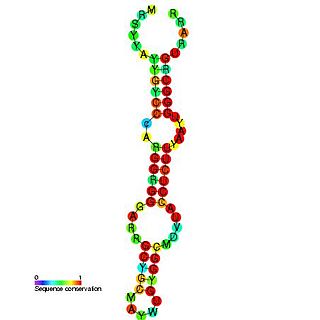
Hepatitis C virus stem-loop VII is a regulatory element found in the coding region of the RNA-dependent RNA polymerase gene, NS5B. Similarly to stem-loop IV, the stem-loop structure is important for colony formation, though its exact function and mechanism are unknown.
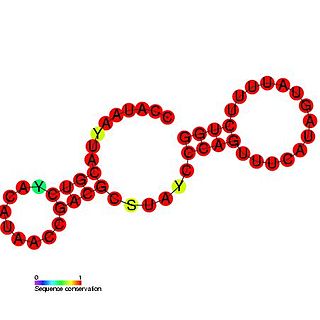
The Potato virus X cis-acting regulatory element is a cis-acting regulatory element found in the 3' UTR of the Potato virus X genome. This element has been found to be required for minus strand RNA accumulation and is essential for efficient viral replication.
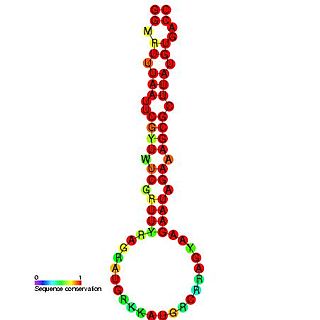
This family represents a rotavirus cis-acting replication element (CRE) found at the 3'-end of rotavirus mRNAs. The family is thought to promote the synthesis of minus strand RNA to form viral dsRNA.
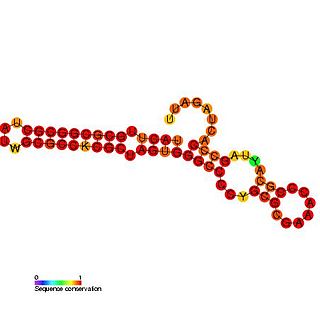
The Rubella virus 3′ cis-acting element RNA family represents a cis-acting element found at the 3′ UTR in the rubella virus. This family contains three conserved step loop structures. Calreticulin (CAL), which is known to bind calcium in most eukaryotic cells, is able to specifically bind to the first stem loop of this RNA. CAL binding is thought to be related to viral pathogenesis and in particular arthritis which occurs frequently in rubella infections in adults and is independent of viral viability. All stem loop structures are thought to be important for efficient viral replication and deletion of stem loop three is known to be lethal.

The Human Parechovirus 1 cis regulatory element is an RNA element which is located in the 5'-terminal 112 nucleotides of the genome of human parechovirus 1 (HPeV1). The element consists of two stem-loop structures together with a pseudoknot. Disruption of any of these elements impairs both viral replication and growth.

Human rhinovirus internal cis-acting regulatory element (CRE) is a CRE from the human rhinoviruses. The CRE is located within the genome segment encoding the capsid proteins so is found in a protein coding region. The element is essential for efficient viral replication and it has been suggested that the CRE is required for initiation of minus-strand RNA synthesis.
Trans-regulatory elements (TRE) are DNA sequences encoding upstream regulators, which may modify or regulate the expression of distant genes. Trans-acting factors interact with cis-regulatory elements to regulate gene expression. TRE mediates expression profiles of a large number of genes via trans-acting factors. While TRE mutations affect gene expression, it is also one of the main driving factors for evolutionary divergence in gene expression.
Cis-acting replication elements bring together the 5′ and 3′ ends during replication of positive-sense single-stranded RNA viruses and double-stranded RNA viruses.
In molecular biology, the Hepatitis A virus cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Hepatitis A virus (HAV). It is larger than the CREs found in related Picornavirus species, but is thought to be functionally similar. It is thought to be involved in uridylylation of VPg.
In molecular biology, the Norovirus cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Norovirus. It occurs near to the 5′ end of the RNA dependant RNA polymerase gene, this is the same location that the Hepatitis A virus cis-acting replication element is found in.
In molecular biology, the Avian encephalitis virus cis-acting replication element (CRE) is an s an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Avian encephalitis virus (AEV). It is structurally similar to the Hepatitis A virus cis-acting replication element.