
Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of genetic recombination, which occurs in the pachytene stage of prophase I of meiosis during a process called synapsis. Synapsis begins before the synaptonemal complex develops and is not completed until near the end of prophase I. Crossover usually occurs when matching regions on matching chromosomes break and then reconnect to the other chromosome.

Genetic recombination is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryotes, genetic recombination during meiosis can lead to a novel set of genetic information that can be further passed on from parents to offspring. Most recombination occurs naturally and can be classified into two types: (1) interchromosomal recombination, occurring through independent assortment of alleles whose loci are on different but homologous chromosomes ; & (2) intrachromosomal recombination, occurring through crossing over.
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be more linked than markers that are far apart. In other words, the nearer two genes are on a chromosome, the lower the chance of recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly unlinked, although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located.

Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands, using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases.

Franklin (Frank) William Stahl is an American molecular biologist and geneticist. With Matthew Meselson, Stahl conducted the famous Meselson-Stahl experiment showing that DNA is replicated by a semiconservative mechanism, meaning that each strand of the DNA serves as a template for production of a new strand.
Gene conversion is the process by which one DNA sequence replaces a homologous sequence such that the sequences become identical after the conversion event. Gene conversion can be either allelic, meaning that one allele of the same gene replaces another allele, or ectopic, meaning that one paralogous DNA sequence converts another.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.
Recombination hotspots are regions in a genome that exhibit elevated rates of recombination relative to a neutral expectation. The recombination rate within hotspots can be hundreds of times that of the surrounding region. Recombination hotspots result from higher DNA break formation in these regions, and apply to both mitotic and meiotic cells. This appellation can refer to recombination events resulting from the uneven distribution of programmed meiotic double-strand breaks.
Mitotic recombination is a type of genetic recombination that may occur in somatic cells during their preparation for mitosis in both sexual and asexual organisms. In asexual organisms, the study of mitotic recombination is one way to understand genetic linkage because it is the only source of recombination within an individual. Additionally, mitotic recombination can result in the expression of recessive alleles in an otherwise heterozygous individual. This expression has important implications for the study of tumorigenesis and lethal recessive alleles. Mitotic homologous recombination occurs mainly between sister chromatids subsequent to replication. Inter-sister homologous recombination is ordinarily genetically silent. During mitosis the incidence of recombination between non-sister homologous chromatids is only about 1% of that between sister chromatids.

Sister chromatid exchange (SCE) is the exchange of genetic material between two identical sister chromatids.

A bivalent is one pair of chromosomes in a tetrad. A tetrad is the association of a pair of homologous chromosomes physically held together by at least one DNA crossover. This physical attachment allows for alignment and segregation of the homologous chromosomes in the first meiotic division. In most organisms, each replicated chromosome elicits formation of DNA double-strand breaks during the leptotene phase. These breaks are repaired by homologous recombination, that uses the homologous chromosome as a template for repair. The search for the homologous target, helped by numerous proteins collectively referred as the synaptonemal complex, cause the two homologs to pair, between the leptotene and the pachytene phases of meiosis I.
Chromosome segregation is the process in eukaryotes by which two sister chromatids formed as a consequence of DNA replication, or paired homologous chromosomes, separate from each other and migrate to opposite poles of the nucleus. This segregation process occurs during both mitosis and meiosis. Chromosome segregation also occurs in prokaryotes. However, in contrast to eukaryotic chromosome segregation, replication and segregation are not temporally separated. Instead segregation occurs progressively following replication.

Bloom syndrome protein is a protein that in humans is encoded by the BLM gene and is not expressed in Bloom syndrome.
DNA topoisomerase 3-alpha is an enzyme that in humans is encoded by the TOP3A gene.
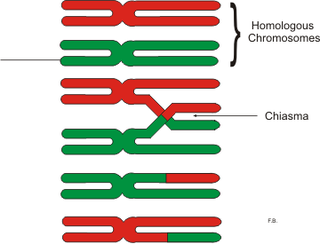
In genetics, a chiasma is the point of contact, the physical link, between two (non-sister) chromatids belonging to homologous chromosomes. At a given chiasma, an exchange of genetic material can occur between both chromatids, what is called a chromosomal crossover, but this is much more frequent during meiosis than mitosis. In meiosis, absence of a chiasma generally results in improper chromosomal segregation and aneuploidy.

Fanconi anemia, complementation group M, also known as FANCM is a human gene. It is an emerging target in cancer therapy, in particular cancers with specific genetic deficiencies.

In genetics, the coefficient of coincidence (c.o.c.) is a measure of interference in the formation of chromosomal crossovers during meiosis. It is generally the case that, if there is a crossover at one spot on a chromosome, this decreases the likelihood of a crossover in a nearby spot. This is called interference.
Rauchvirus is a genus of viruses in the order Caudovirales, in the family Podoviridae. Bacteria serve as natural hosts. The genus contains only one species: Bordetella virus BPP1.

PR domain zinc finger protein 9 is a protein that in humans is encoded by the PRDM9 gene. PRDM9 is responsible for positioning recombination hotspots during meiosis by binding a DNA sequence motif encoded in its zinc finger domain. PRDM9 is the only speciation gene found so far in mammals, and is one of the fastest evolving genes in the genome.

Synthesis-dependent strand annealing (SDSA) is a major mechanism of homology-directed repair of DNA double-strand breaks (DSBs). Although many of the features of SDSA were first suggested in 1976, the double-Holliday junction model proposed in 1983 was favored by many researchers. In 1994, studies of double-strand gap repair in Drosophila were found to be incompatible with the double-Holliday junction model, leading researchers to propose a model they called synthesis-dependent strand annealing. Subsequent studies of meiotic recombination in S. cerevisiae found that non-crossover products appear earlier than double-Holliday junctions or crossover products, challenging the previous notion that both crossover and non-crossover products are produced by double-Holliday junctions and leading the authors to propose that non-crossover products are generated through SDSA.













