In genetics and biochemistry, sequencing means to determine the primary structure of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which succinctly summarizes much of the atomic-level structure of the sequenced molecule.

Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication.
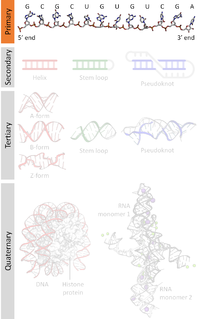
A nucleic acid sequence is a succession of bases signified by a series of a set of five different letters that indicate the order of nucleotides forming alleles within a DNA or RNA (GACU) molecule. By convention, sequences are usually presented from the 5' end to the 3' end. For DNA, the sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the primary structure.
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functional elements such as regulatory regions. Gene finding is one of the first and most important steps in understanding the genome of a species once it has been sequenced.
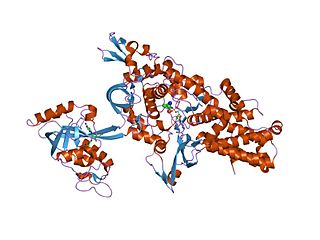
An aminoacyl-tRNA synthetase, also called tRNA-ligase, is an enzyme that attaches the appropriate amino acid onto its corresponding tRNA. It does so by catalyzing the transesterification of a specific cognate amino acid or its precursor to one of all its compatible cognate tRNAs to form an aminoacyl-tRNA. In humans, the 20 different types of aa-tRNA are made by the 20 different aminoacyl-tRNA synthetases, one for each amino acid of the genetic code.

In biology, a gene is a basic unit of heredity and a sequence of nucleotides in DNA that encodes the synthesis of a gene product, either RNA or protein.

RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology and evolutionary biology. It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see Glossary of evolutionary biology.
The Reference Sequence (RefSeq) database is an open access, annotated and curated collection of publicly available nucleotide sequences and their protein products. RefSeq was first introduced in 2000. This database is built by National Center for Biotechnology Information (NCBI), and, unlike GenBank, provides only a single record for each natural biological molecule for major organisms ranging from viruses to bacteria to eukaryotes.

A wide variety of non-coding RNAs have been identified in various species of organisms known to science. However, RNAs have also been identified in "metagenomics" sequences derived from samples of DNA or RNA extracted from the environment, which contain unknown species. Initial work in this area detected homologs of known bacterial RNAs in such metagenome samples. Many of these RNA sequences were distinct from sequences within cultivated bacteria, and provide the potential for additional information on the RNA classes to which they belong.

The glutamine riboswitch is a conserved RNA structure that was predicted by bioinformatics. It is present in a variety of lineages of cyanobacteria, as well as some phages that infect cyanobacteria. It is also found in DNA extracted from uncultivated bacteria living in the ocean that are presumably species of cyanobacteria.

The potC RNA motif is a conserved RNA structure discovered using bioinformatics. The RNA is detected only in genome sequences derived from DNA that was extracted from uncultivated marine bacteria. Thus, this RNA is present in environmental samples, but not yet found in any cultivated organism. potC RNAs are located in the presumed 5' untranslated regions of genes predicted to encode either membrane transport proteins or peroxiredoxins. Therefore, it was hypothesized that potC RNAs are cis-regulatory elements, but their detailed function is unknown.

The Termite-leu RNA motif is a conserved RNA structure discovered by bioinformatics. It is found only in DNA sequences extracted from uncultivated bacteria living in termite hindguts, and has not yet been detected in any known cultivated organism. In many cases, Termite-leu RNAs are found in the likely 5′ untranslated regions of multive genes related to the synthesis of the amino acid leucine. However, in several cases it is not found in this type of location. Therefore, it was considered ambiguous as to whether Termite-leu RNAs constitute cis-regulatory elements.

The Whalefall-1 RNA motif refers to a conserved RNA structure that was discovered using bioinformatics. Structurally, the motif consists of two stem-loops, the second of which is often terminated by a CUUG tetraloop, which is an energetically favorable RNA sequence. Whalefall-1 RNAs are found only in DNA extracted from uncultivated bacteria found on whale fall, i.e., a whale carcass. As of 2010, Whalefall-1 RNAs have not been detected in any known, cultivated species of bacteria, and are thus one of several RNAs present in environmental samples.
The Ocean-V RNA motif is a conserved RNA structure discovered using bioinformatics. Only a few Ocean-V RNA sequences have been detected, all in sequences derived from DNA that was extracted from uncultivated bacteria found in ocean water. As of 2010, no Ocean-V RNA has been detected in any known, cultivated organism.
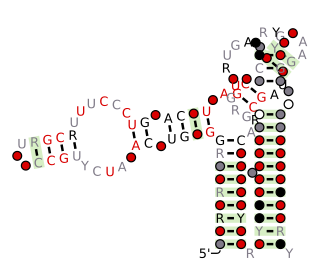
The Flavobacterium-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Flavobacterium-1 motif RNAs are found in metagenomic samples from the environment, and only one example of this motif is present in a classified organism. This organism is Flavobacterium sp. SCGC AAA160-P02, which belongs to the bacterial phylum Bacteroidetes.

The Latescibacteria, OD1, OP11, TM7 RNA motif is a conserved RNA structure that was discovered by bioinformatics. LOOT motif RNAs are found in multiple bacterial phyla that have only recently been discovered, and are currently not well understood: Latescibacteria, OD1/Parcubacteria, OP11 AND TM7. In some cases, no specific organism has been isolated in the relevant phylum, but the existence of the bacterial phylum is known only through analysis of metagenomic sequences. Curiously, the LOOT motif is not known in any phylum that has been studied for a long time.

The Mu-like gpT Downstream Element RNA motif is a conserved RNA structure that was discovered by bioinformatics. The Mu-gpT-DE motif is only found in metagenomic sequences arising from unknown organisms.

Monodnaviria is a realm of viruses that includes all single-stranded DNA viruses that encode an endonuclease of the HUH superfamily that initiates rolling circle replication of the circular viral genome. Viruses descended from such viruses are also included in the realm, including certain linear single-stranded DNA (ssDNA) viruses and circular double-stranded DNA (dsDNA) viruses. These atypical members typically replicate through means other than rolling circle replication.
Retrozymes are a family of retrotransposons first discovered in the genomes of plants but now also known in genomes of animals. Retrozymes contain a hammerhead ribozyme (HHR) in their sequences, although they do not possess any coding regions. Retrozymes are nonautonomous retroelements, and so borrow proteins from other elements to move into new regions of a genome. Retrozymes are actively transcribed into covalently closed circular RNAs and are detected in both polarities, which may indicate the use of rolling circle replication in their lifecycle.














