
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993.
Protein engineering is the process of developing useful or valuable proteins through the design and production of unnatural polypeptides, often by altering amino acid sequences found in nature. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It has been used to improve the function of many enzymes for industrial catalysis. It is also a product and services market, with an estimated value of $168 billion by 2017.

Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA and amplification of specific DNA targets using polymerase chain reaction (PCR). It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Confusion can arise because some authors use the acronym RT-PCR to denote real-time PCR. In this article, RT-PCR will denote Reverse Transcription PCR. Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings.
The touchdown polymerase chain reaction or touchdown style polymerase chain reaction is a method of polymerase chain reaction by which primers avoid amplifying nonspecific sequences. The annealing temperature during a polymerase chain reaction determines the specificity of primer annealing. The melting point of the primer sets the upper limit on annealing temperature. At temperatures just above this point, only very specific base pairing between the primer and the template will occur. At lower temperatures, the primers bind less specifically. Nonspecific primer binding obscures polymerase chain reaction results, as the nonspecific sequences to which primers anneal in early steps of amplification will "swamp out" any specific sequences because of the exponential nature of polymerase amplification.
In molecular biology, an amplicon is a piece of DNA or RNA that is the source and/or product of amplification or replication events. It can be formed artificially, using various methods including polymerase chain reactions (PCR) or ligase chain reactions (LCR), or naturally through gene duplication. In this context, amplification refers to the production of one or more copies of a genetic fragment or target sequence, specifically the amplicon. As it refers to the product of an amplification reaction, amplicon is used interchangeably with common laboratory terms, such as "PCR product."
Cycling probe technology (CPT) is a molecular biological technique for detecting specific DNA sequences. CPT operates under isothermal conditions. In some applications, CPT offers an alternative to PCR. However, unlike PCR, CPT does not generate multiple copies of the target DNA itself, and the amplification of the signal is linear, in contrast to the exponential amplification of the target DNA in PCR. CPT uses a sequence specific chimeric probe which hybridizes to a complementary target DNA sequence and becomes a substrate for RNase H. Cleavage occurs at the RNA internucleotide linkages and results in dissociation of the probe from the target, thereby making it available for the next probe molecule. Integrated electrokinetic systems have been developed for use in CPT.
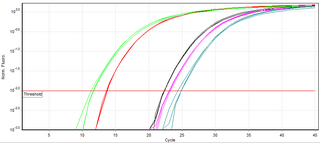
A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively.
TaqMan probes are hydrolysis probes that are designed to increase the specificity of quantitative PCR. The method was first reported in 1991 by researcher Kary Mullis at Cetus Corporation, and the technology was subsequently developed by Hoffmann-La Roche for diagnostic assays and by Applied Biosystems for research applications.
Multiplex ligation-dependent probe amplification (MLPA) is a variation of the multiplex polymerase chain reaction that permits amplification of multiple targets with only a single primer pair. It detects copy number changes at the molecular level, and software programs are used for analysis. Identification of deletions or duplications can indicate pathogenic mutations, thus MLPA is an important diagnostic tool used in clinical pathology laboratories worldwide.

Bisulfitesequencing (also known as bisulphite sequencing) is the use of bisulfite treatment of DNA before routine sequencing to determine the pattern of methylation. DNA methylation was the first discovered epigenetic mark, and remains the most studied. In animals it predominantly involves the addition of a methyl group to the carbon-5 position of cytosine residues of the dinucleotide CpG, and is implicated in repression of transcriptional activity.
Digital polymerase chain reaction is a biotechnological refinement of conventional polymerase chain reaction methods that can be used to directly quantify and clonally amplify nucleic acids strands including DNA, cDNA, or RNA. The key difference between dPCR and qPCR lies in the method of measuring nucleic acids amounts, with the former being a more precise method than PCR, though also more prone to error in the hands of inexperienced users. PCR carries out one reaction per single sample. dPCR also carries out a single reaction within a sample, however the sample is separated into a large number of partitions and the reaction is carried out in each partition individually. This separation allows a more reliable collection and sensitive measurement of nucleic acid amounts. The method has been demonstrated as useful for studying variations in gene sequences—such as copy number variants and point mutations.
Melting curve analysis is an assessment of the dissociation characteristics of double-stranded DNA during heating. As the temperature is raised, the double strand begins to dissociate leading to a rise in the absorbance intensity, hyperchromicity. The temperature at which 50% of DNA is denatured is known as the melting temperature. Measurement of melting temperature can help us predict species by just studying the melting temperature. This is because every organism has a specific melting curve.
The versatility of polymerase chain reaction (PCR) has led to modifications of the basic protocol being used in a large number of variant techniques designed for various purposes. This article summarizes many of the most common variations currently or formerly used in molecular biology laboratories; familiarity with the fundamental premise by which PCR works and corresponding terms and concepts is necessary for understanding these variant techniques.
High Resolution Melt (HRM) analysis is a powerful technique in molecular biology for the detection of mutations, polymorphisms and epigenetic differences in double-stranded DNA samples. It was discovered and developed by Idaho Technology and the University of Utah. It has advantages over other genotyping technologies, namely:
A primer dimer (PD) is a potential by-product in the polymerase chain reaction (PCR), a common biotechnological method. As its name implies, a PD consists of two primer molecules that have attached (hybridized) to each other because of strings of complementary bases in the primers. As a result, the DNA polymerase amplifies the PD, leading to competition for PCR reagents, thus potentially inhibiting amplification of the DNA sequence targeted for PCR amplification. In quantitative PCR, PDs may interfere with accurate quantification.
Multiplex polymerase chain reaction refers to the use of polymerase chain reaction to amplify several different DNA sequences simultaneously. This process amplifies DNA in samples using multiple primers and a temperature-mediated DNA polymerase in a thermal cycler. The primer design for all primers pairs has to be optimized so that all primer pairs can work at the same annealing temperature during PCR.
A quantitative PCR instrument is a machine that amplifies and detects DNA. It combines the functions of a thermal cycler and a fluorimeter, enabling the process of quantitative PCR.
The terms "relative fluorescence units" (RFU) and "RFU peak" refer to measurements in electrophoresis methods, such as for DNA analysis. A "relative fluorescence unit" is a unit of measurement used in analysis which employs fluorescence detection. Fluorescence is detected using a charge-coupled device (CCD) array, when the labeled fragments, which are separated within a capillary by using electrophoresis, are energized by laser light and travel across the detection window. A computer program measures the results, determining the quantity or size of the fragments, at each data point, from the level of fluorescence intensity. Samples which contain higher quantities of amplified DNA will have higher corresponding RFU values.
In biotechnology BEAMing, which stands for beads, emulsion, amplification, magnetics, is a highly sensitive digital PCR method that combines emulsion PCR and flow cytometry to identify and quantify specific somatic mutations present in DNA.
The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines are a set of protocols for conducting and reporting quantitative real-time PCR experiments and data, as devised by Bustin et al. in 2009. They were devised after a paper was published in 2002 that claimed to detect measles virus in children with autism through the use of RT-qPCR, but the results proved to be completely unreproducible by other scientists. The authors themselves also did not try to reproduce them and the raw data was found to have a large amount of errors and basic mistakes in analysis. This incident prompted Stephen Bustin to create the MIQE guidelines to provide a baseline level of quality for qPCR data published in scientific literature.




