Related Research Articles

Lissencephaly is a set of rare brain disorders whereby the whole or parts of the surface of the brain appear smooth. It is caused by defective neuronal migration during the 12th to 24th weeks of gestation resulting in a lack of development of brain folds (gyri) and grooves (sulci). It is a form of cephalic disorder. Terms such as agyria and pachygyria are used to describe the appearance of the surface of the brain.

In genetics, a deletion is a mutation in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur, which result in the deletion of a part of the chromosome. The breaks can be induced by heat, viruses, radiation, or chemical reactions. When a chromosome breaks, if a part of it is deleted or lost, the missing piece of chromosome is referred to as a deletion or a deficiency.
Miller–Dieker syndrome, Miller–Dieker lissencephaly syndrome (MDLS), and chromosome 17p13.3 deletion syndrome is a micro deletion syndrome characterized by congenital malformations. Congenital malformations are physical defects detectable in an infant at birth which can involve many different parts of the body including the brain, hearts, lungs, liver, bones, or intestinal tract. MDS is a contiguous gene syndrome – a disorder due to the deletion of multiple gene loci adjacent to one another. The disorder arises from the deletion of part of the small arm of chromosome 17p, leading to partial monosomy. There may be unbalanced translocations, or the presence of a ring chromosome 17.

Loss of heterozygosity (LOH) is a type of genetic abnormality in diploid organisms in which one copy of an entire gene and its surrounding chromosomal region are lost. Since diploid cells have two copies of their genes, one from each parent, a single copy of the lost gene still remains when this happens, but any heterozygosity is no longer present.

Chromosome 22 is one of the 23 pairs of chromosomes in human cells. Humans normally have two copies of chromosome 22 in each cell. Chromosome 22 is the second smallest human chromosome, spanning about 51 million DNA base pairs and representing between 1.5 and 2% of the total DNA in cells.
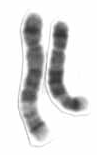
Chromosome 5 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 5 spans about 182 million base pairs and represents almost 6% of the total DNA in cells. Chromosome 5 is the 5th largest human chromosome, yet has one of the lowest gene densities. This is partially explained by numerous gene-poor regions that display a remarkable degree of non-coding and syntenic conservation with non-mammalian vertebrates, suggesting they are functionally constrained.

Chromosome 15 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 15 spans about 99.7 million base pairs and represents between 3% and 3.5% of the total DNA in cells. Chromosome 15 is an acrocentric chromosome, with a very small short arm, which contains few protein coding genes among its 19 million base pairs. It has a larger long arm that is gene rich, spanning about 83 million base pairs.
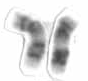
Chromosome 17 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 17 spans more than 84 million base pairs and represents between 2.5 and 3% of the total DNA in cells.

T-box transcription factor TBX1 also known as T-box protein 1 and testis-specific T-box protein is a protein that in humans is encoded by the TBX1 gene. Genes in the T-box family are transcription factors that play important roles in the formation of tissues and organs during embryonic development. To carry out these roles, proteins made by this gene family bind to specific areas of DNA called T-box binding element (TBE) to control the expression of target genes.
The short-stature homeobox gene (SHOX), also known as short-stature-homeobox-containing gene, is a gene located on both the X and Y chromosomes, which is associated with short stature in humans if mutated or present in only one copy (haploinsufficiency).

22q13 deletion syndrome, known as Phelan–McDermid syndrome (PMS), is a genetic disorder caused by deletions or rearrangements on the q terminal end of chromosome 22. Any abnormal genetic variation in the q13 region that presents with significant manifestations (phenotype) typical of a terminal deletion may be diagnosed as 22q13 deletion syndrome. There is disagreement among researchers as to the exact definition of 22q13 deletion syndrome. The Developmental Synaptopathies Consortium defines PMS as being caused by SHANK3 mutations, a definition that appears to exclude terminal deletions. The requirement to include SHANK3 in the definition is supported by many but not by those who first described 22q13 deletion syndrome.
A chromosomal abnormality, chromosomal anomaly, chromosomal aberration, chromosomal mutation, or chromosomal disorder is a missing, extra, or irregular portion of chromosomal DNA. These can occur in the form of numerical abnormalities, where there is an atypical number of chromosomes, or as structural abnormalities, where one or more individual chromosomes are altered. Chromosome mutation was formerly used in a strict sense to mean a change in a chromosomal segment, involving more than one gene. Chromosome anomalies usually occur when there is an error in cell division following meiosis or mitosis. Chromosome abnormalities may be detected or confirmed by comparing an individual's karyotype, or full set of chromosomes, to a typical karyotype for the species via genetic testing.
Uner Tan syndrome (UTS) is a syndrome that was discovered by the Turkish evolutionary biologist Üner Tan. People affected by UTS walk with a quadrupedal locomotion and often have severe learning disabilities. Tan postulated that this is an example of "reverse evolution" (atavism). The proposed syndrome was featured in the 2006 BBC2 documentary The Family That Walks On All Fours.

Hypermethylated in cancer 1 protein is a protein that in humans is encoded by the HIC1 gene.

Developmentally-regulated GTP-binding protein 2 is a protein that in humans is encoded by the DRG2 gene.

Diphthamide biosynthesis protein 1 is a protein that in humans is encoded by the DPH1 gene. It encodes a protein that performs posttranslational modification of histidine-715 on eukaryotic translation elongation factor 2 to diphthamide. This modification appears to be important in the translation of Cyclin D in ovarian cells. DPH1 is mutated in 90% of ovarian cancers end stage, usually by loss of heterozygosity.

Glyoxalase domain-containing protein 4 is an enzyme that in humans is encoded by the GLOD4 gene.
1q21.1 deletion syndrome is a rare aberration of chromosome 1. A human cell has one pair of identical chromosomes on chromosome 1. With the 1q21.1 deletion syndrome, one chromosome of the pair is not complete, because a part of the sequence of the chromosome is missing. One chromosome has the normal length and the other is too short.

A microdeletion syndrome is a syndrome caused by a chromosomal deletion smaller than 5 million base pairs spanning several genes that is too small to be detected by conventional cytogenetic methods or high resolution karyotyping. Detection is done by fluorescence in situ hybridization (FISH). Larger chromosomal deletion syndromes are detectable using karyotyping techniques.

DiGeorge syndrome, also known as 22q11.2 deletion syndrome, is a syndrome caused by a microdeletion on the long arm of chromosome 22. While the symptoms can vary, they often include congenital heart problems, specific facial features, frequent infections, developmental disability, intellectual disability and cleft palate. Associated conditions include kidney problems, schizophrenia, hearing loss and autoimmune disorders such as rheumatoid arthritis or Graves' disease.
References
- ↑ "Entrez Gene: Chromosome 17p13.1 deletion syndrome".
- ↑ Carvalho CM, Vasanth S, Shinawi M, Russell C, Ramocki MB, Brown CW, et al. (November 2014). "Dosage changes of a segment at 17p13.1 lead to intellectual disability and microcephaly as a result of complex genetic interaction of multiple genes". American Journal of Human Genetics. 95 (5): 565–78. doi:10.1016/j.ajhg.2014.10.006. PMC 4225592 . PMID 25439725.