
A Southern blot is a method used in molecular biology for detection of a specific DNA sequence in DNA samples. Southern blotting combines transfer of electrophoresis-separated DNA fragments to a filter membrane and subsequent fragment detection by probe hybridization.

In biochemistry, allosteric regulation is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site.
A biosensor is an analytical device, used for the detection of a chemical substance, that combines a biological component with a physicochemical detector. The sensitive biological element, e.g. tissue, microorganisms, organelles, cell receptors, enzymes, antibodies, nucleic acids, etc., is a biologically derived material or biomimetic component that interacts with, binds with, or recognizes the analyte under study. The biologically sensitive elements can also be created by biological engineering. The transducer or the detector element, which transforms one signal into another one, works in a physicochemical way: optical, piezoelectric, electrochemical, electrochemiluminescence etc., resulting from the interaction of the analyte with the biological element, to easily measure and quantify. The biosensor reader device connects with the associated electronics or signal processors that are primarily responsible for the display of the results in a user-friendly way. This sometimes accounts for the most expensive part of the sensor device, however it is possible to generate a user friendly display that includes transducer and sensitive element. The readers are usually custom-designed and manufactured to suit the different working principles of biosensors.
Nanosensors are nanoscale devices that measure physical quantities and convert these to signals that can be detected and analyzed. There are several ways proposed today to make nanosensors; these include top-down lithography, bottom-up assembly, and molecular self-assembly. There are different types of nanosensors in the market and in development for various applications, most notably in defense, environmental, and healthcare industries. These sensors share the same basic workflow: a selective binding of an analyte, signal generation from the interaction of the nanosensor with the bio-element, and processing of the signal into useful metrics.

A nanopore is a pore of nanometer size. It may, for example, be created by a pore-forming protein or as a hole in synthetic materials such as silicon or graphene.

Nanopore sequencing is a third generation approach used in the sequencing of biopolymers- specifically, polynucleotides in the form of DNA or RNA.

Fluorescence in situ hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only those parts of a nucleic acid sequence with a high degree of sequence complementarity. It was developed by biomedical researchers in the early 1980s to detect and localize the presence or absence of specific DNA sequences on chromosomes. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling, medicine, and species identification. FISH can also be used to detect and localize specific RNA targets in cells, circulating tumor cells, and tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression within cells and tissues.
Aptamers are oligonucleotide or peptide molecules that bind to a specific target molecule. Aptamers are usually created by selecting them from a large random sequence pool, but natural aptamers also exist in riboswitches. Aptamers can be used for both basic research and clinical purposes as macromolecular drugs. Aptamers can be combined with ribozymes to self-cleave in the presence of their target molecule. These compound molecules have additional research, industrial and clinical applications.

An ion-sensitive field-effect transistor (ISFET) is a field-effect transistor used for measuring ion concentrations in solution; when the ion concentration (such as H+, see pH scale) changes, the current through the transistor will change accordingly. Here, the solution is used as the gate electrode. A voltage between substrate and oxide surfaces arises due to an ion sheath. It is a special type of MOSFET (metal-oxide-semiconductor field-effect transistor), and shares the same basic structure, but with the metal gate replaced by an ion-sensitive membrane, electrolyte solution and reference electrode. Invented in 1970, the ISFET was the first biosensor FET (BioFET).

In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ligare, which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure. The instance of binding occurs over an infinitesimal range of time and space, so the rate constant is usually a very small number.
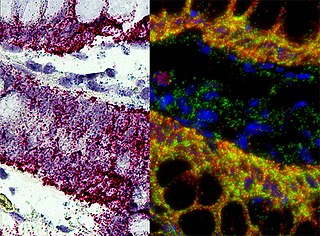
In situ hybridization (ISH) is a type of hybridization that uses a labeled complementary DNA, RNA or modified nucleic acids strand to localize a specific DNA or RNA sequence in a portion or section of tissue or if the tissue is small enough, in the entire tissue, in cells, and in circulating tumor cells (CTCs). This is distinct from immunohistochemistry, which usually localizes proteins in tissue sections.
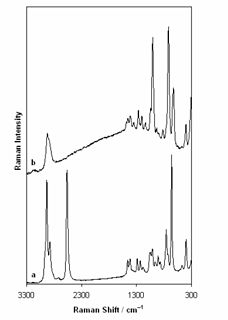
Surface-enhanced Raman spectroscopy or surface-enhanced Raman scattering (SERS) is a surface-sensitive technique that enhances Raman scattering by molecules adsorbed on rough metal surfaces or by nanostructures such as plasmonic-magnetic silica nanotubes. The enhancement factor can be as much as 1010 to 1011, which means the technique may detect single molecules.

Nanofluidics is the study of the behavior, manipulation, and control of fluids that are confined to structures of nanometer characteristic dimensions. Fluids confined in these structures exhibit physical behaviors not observed in larger structures, such as those of micrometer dimensions and above, because the characteristic physical scaling lengths of the fluid, very closely coincide with the dimensions of the nanostructure itself.
A biointerface is the region of contact between a biomolecule, cell, biological tissue or living organism or organic material considered living with another biomaterial or inorganic/organic material. The motivation for biointerface science stems from the urgent need to increase the understanding of interactions between biomolecules and surfaces. The behavior of complex macromolecular systems at materials interfaces are important in the fields of biology, biotechnology, diagnostics, and medicine. Biointerface science is a multidisciplinary field in which biochemists who synthesize novel classes of biomolecules cooperate with scientists who have developed the tools to position biomolecules with molecular precision, scientists who have developed new spectroscopic techniques to interrogate these molecules at the solid-liquid interface, and people who integrate these into functional devices.
Nanofluidic circuitry is a nanotechnology aiming for control of fluids in nanometer scale. Due to the effect of an electrical double layer within the fluid channel, the behavior of nanofluid is observed to be significantly different compared with its microfluidic counterparts. Its typical characteristic dimensions fall within the range of 1–100 nm. At least one dimension of the structure is in nanoscopic scale. Phenomena of fluids in nano-scale structure are discovered to be of different properties in electrochemistry and fluid dynamics.

Silicene is a two-dimensional allotrope of silicon, with a hexagonal honeycomb structure similar to that of graphene. Contrary to graphene, silicene is not flat, but has a periodically buckled topology; the coupling between layers in silicene is much stronger than in multilayered graphene; and the oxidized form of silicene, 2D silica, has a very different chemical structure from graphene oxide.

Spherical nucleic acids (SNAs) are nanostructures that consist of a densely packed, highly oriented arrangement of linear nucleic acids in a three-dimensional, spherical geometry. This novel three-dimensional architecture is responsible for many of the SNA's novel chemical, biological, and physical properties that make it useful in biomedicine and materials synthesis. SNAs were first introduced in 1996 by Chad Mirkin’s group at Northwestern University.
Potential graphene applications include lightweight, thin, and flexible electric/photonics circuits, solar cells, and various medical, chemical and industrial processes enhanced or enabled by the use of new graphene materials.
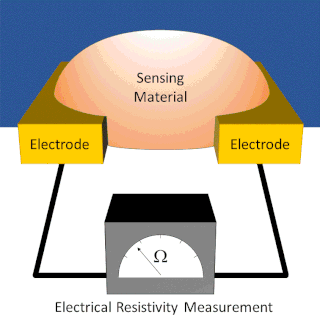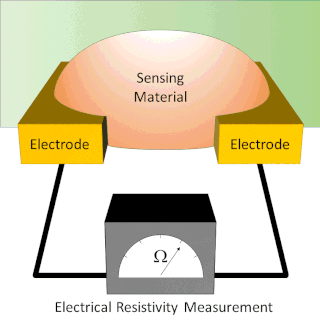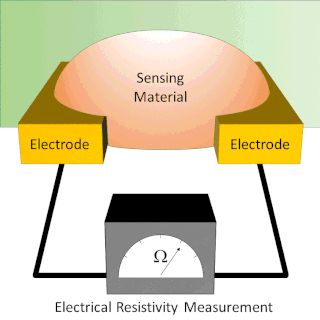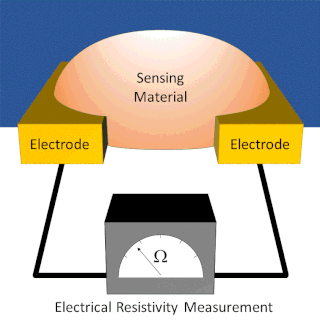
A chemiresistor is a material that changes its electrical resistance in response to changes in the nearby chemical environment. Chemiresistors are a class of chemical sensors that rely on the direct chemical interaction between the sensing material and the analyte. The sensing material and the analyte can interact by covalent bonding, hydrogen bonding, or molecular recognition. Several different materials have chemiresistor properties: metal-oxide semiconductors, some conductive polymers, and nanomaterials like graphene, carbon nanotubes and nanoparticles. Typically these materials are used as partially selective sensors in devices like electronic tongues or electronic noses.

A field-effect transistor-based biosensor, also known as a biosensor field-effect transistor, field-effect biosensor (FEB), or biosensor MOSFET, is a field-effect transistor that is gated by changes in the surface potential induced by the binding of molecules. When charged molecules, such as biomolecules, bind to the FET gate, which is usually a dielectric material, they can change the charge distribution of the underlying semiconductor material resulting in a change in conductance of the FET channel. A Bio-FET consists of two main compartments: one is the biological recognition element and the other is the field-effect transistor. The BioFET structure is largely based on the ion-sensitive field-effect transistor (ISFET), a type of metal-oxide-semiconductor field-effect transistor (MOSFET) where the metal gate is replaced by an ion-sensitive membrane, electrolyte solution and reference electrode.














