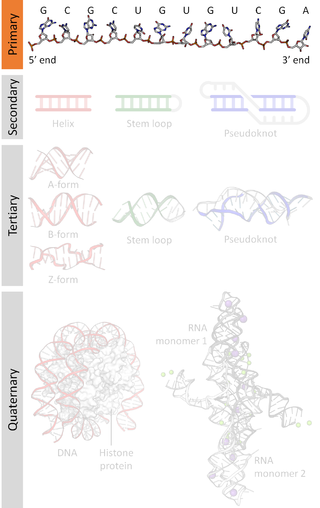
A nucleic acid sequence is a succession of bases within the nucleotides forming alleles within a DNA or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the order of the nucleotides. By convention, sequences are usually presented from the 5' end to the 3' end. For DNA, with its double helix, there are two possible directions for the notated sequence; of these two, the sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the primary structure.
Integrons are genetic mechanisms that allow bacteria to adapt and evolve rapidly through the stockpiling and expression of new genes. These genes are embedded in a specific genetic structure called gene cassette that generally carries one promoterless open reading frame (ORF) together with a recombination site (attC). Integron cassettes are incorporated to the attI site of the integron platform by site-specific recombination reactions mediated by the integrase.

Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as a list of bases which are paired in a nucleic acid molecule. The secondary structures of biological DNAs and RNAs tend to be different: biological DNA mostly exists as fully base paired double helices, while biological RNA is single stranded and often forms complex and intricate base-pairing interactions due to its increased ability to form hydrogen bonds stemming from the extra hydroxyl group in the ribose sugar.

The ARRPOF RNA motif is a conserved RNA structure that was discovered by bioinformatics.

The cold-seep-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics.

The DUF2815 RNA motif is a conserved RNA structure that was discovered by bioinformatics. As of 2018, the DUF2815 motif has not been identified in any classified organism, but is known through metagenomic sequences isolated from environmental sources.
The freshwater-2 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Freshwater-2 motif RNAs are found in metagenomic sequences that are isolated from aquatic and especially freshwater environments. As of 2018, no freshwater-2 RNA has been identified in a classified organism.

The FTHFS RNA motif is a conserved RNA structure that was discovered by bioinformatics. FTHFS motifs are found in metagenomic sequences derived from samples of the human gut.

The int-alpA RNA motif is a conserved RNA structure that was discovered by bioinformatics. int-alpA motif RNAs are found in Pseudomonadota, and one example is known in each of Acidobacteriota and Planctomycetota. An int-alpA was also detected in a purified phage, specifically Thalssomonas phage BA3.
IS605-orfB RNA motifs refer to conserved RNA or DNA structures that were discovered by bioinformatics. Although such motifs were published as a RNA candidates, there is some reason to suspect that they might function as a single-stranded DNA. In terms of secondary structure, RNA and DNA are difficult to distinguish when only sequence information is available. If the motifs function as RNA, they likely are small RNAs, that are independently transcribed.

The PGK RNA motif is a conserved RNA structure that was discovered by bioinformatics. PGK motif RNAs are found in metagenomic sequences isolated from the gastrointestinal tract of mammals. PGK RNAs have not yet been detected in a classified organism.

The poplar-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. As of 2018, all known examples of the poplar-1 motif are found in metagenomic sequences; no poplar-1 RNA has yet been found in a classified organism. Poplar-1 RNAs were particularly common in a metagenomic sample from decaying yellow poplar tree wood chips.

The Poribacteria-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. The Poribacteria-1 motif is found only in the candidate bacterial phylum known as Poribacteria, and all 6 Poribacteria-1 RNAs are actually found in one organism, Candidatus Poribacteria sp. WGA-4E. All but one of these RNAs occur within roughly 6 kilobases of genomic DNA, and each of the 5 RNAs occurs between a different pair of protein-coding genes. This arrangement could suggest that the motif functions on the level of single-stranded DNA as attC sites that are part of an integron. It is also possible that Poribacteria-1 RNAs are cis-regulatory elements that regulate genes that happen to often be nearby to one another, or that the RNAs function in trans as small RNAs.
The sbcC RNA motif is a conserved RNA structure that was discovered by bioinformatics. The sbcC motif has, as of 2018, only been detected in metagenomic sequences, and the identities of organisms that contain these RNAs is unknown.

The ssNA-helicase RNA motif is a conserved RNA structure that was discovered by bioinformatics. Although the ssNA-helicase motif was published as an RNA candidate, there is some reason to suspect that it might function as a single-stranded DNA. In terms of secondary structure, RNA and DNA are difficult to distinguish when only sequence information is available.

The Staphylococcus-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. A Staphylococcus-1 motif RNAs is found in Staphylococcus species CAG-324, which has not yet been more precisely classified. Other examples of Staphylococcus-1 RNAs are present in metagenomic sequences that do not correspond to a classified organism. It is assumed that the organism corresponding to these sequences are related to the Staphylococcus species.
The throat-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. The throat-1 motif has not yet been found in a classified organism, but rather is known only from metagenomic sequences isolated from the human throat is, more rarely, tongue.

The xerDC RNA motif is a conserved RNA structure that was discovered by bioinformatics. xerDC motif RNAs are found in Clostridia.
This glossary of cellular and molecular biology is a list of definitions of terms and concepts commonly used in the study of cell biology, molecular biology, and related disciplines, including molecular genetics, biochemistry, and microbiology. It is split across two articles:
This glossary of cellular and molecular biology is a list of definitions of terms and concepts commonly used in the study of cell biology, molecular biology, and related disciplines, including genetics, biochemistry, and microbiology. It is split across two articles:













