
Microfluidics refers to a system that manipulates a small amount of fluids using small channels with sizes ten to hundreds micrometres. It is a multidisciplinary field that involves molecular analysis, molecular biology, and microelectronics. It has practical applications in the design of systems that process low volumes of fluids to achieve multiplexing, automation, and high-throughput screening. Microfluidics emerged in the beginning of the 1980s and is used in the development of inkjet printheads, DNA chips, lab-on-a-chip technology, micro-propulsion, and micro-thermal technologies.

Digital microfluidics (DMF) is a platform for lab-on-a-chip systems that is based upon the manipulation of microdroplets. Droplets are dispensed, moved, stored, mixed, reacted, or analyzed on a platform with a set of insulated electrodes. Digital microfluidics can be used together with analytical analysis procedures such as mass spectrometry, colorimetry, electrochemical, and electrochemiluminescense.
A lab-on-a-chip (LOC) is a device that integrates one or several laboratory functions on a single integrated circuit of only millimeters to a few square centimeters to achieve automation and high-throughput screening. LOCs can handle extremely small fluid volumes down to less than pico-liters. Lab-on-a-chip devices are a subset of microelectromechanical systems (MEMS) devices and sometimes called "micro total analysis systems" (μTAS). LOCs may use microfluidics, the physics, manipulation and study of minute amounts of fluids. However, strictly regarded "lab-on-a-chip" indicates generally the scaling of single or multiple lab processes down to chip-format, whereas "μTAS" is dedicated to the integration of the total sequence of lab processes to perform chemical analysis.

DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.
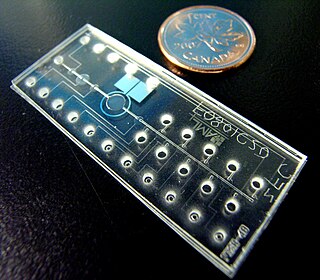
Bio-MEMS is an abbreviation for biomedical microelectromechanical systems. Bio-MEMS have considerable overlap, and is sometimes considered synonymous, with lab-on-a-chip (LOC) and micro total analysis systems (μTAS). Bio-MEMS is typically more focused on mechanical parts and microfabrication technologies made suitable for biological applications. On the other hand, lab-on-a-chip is concerned with miniaturization and integration of laboratory processes and experiments into single chips. In this definition, lab-on-a-chip devices do not strictly have biological applications, although most do or are amenable to be adapted for biological purposes. Similarly, micro total analysis systems may not have biological applications in mind, and are usually dedicated to chemical analysis. A broad definition for bio-MEMS can be used to refer to the science and technology of operating at the microscale for biological and biomedical applications, which may or may not include any electronic or mechanical functions. The interdisciplinary nature of bio-MEMS combines material sciences, clinical sciences, medicine, surgery, electrical engineering, mechanical engineering, optical engineering, chemical engineering, and biomedical engineering. Some of its major applications include genomics, proteomics, molecular diagnostics, point-of-care diagnostics, tissue engineering, single cell analysis and implantable microdevices.

RNA-Seq is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome.
Cell sorting is the process through which a particular cell type is separated from others contained in a sample on the basis of its physical or biological properties, such as size, morphological parameters, viability and both extracellular and intracellular protein expression. The homogeneous cell population obtained after sorting can be used for a variety of applications including research, diagnosis, and therapy.
Integromics was a global bioinformatics company headquartered in Granada, Spain and Madrid. The company had subsidiaries in the United States and United Kingdom, and distributors in 10 countries. Integromics specialised in bioinformatics software for data management and data analysis in genomics and proteomics. The company provided a line of products that serve gene expression, sequencing, and proteomics markets. Customers included genomic research centers, pharmaceutical companies, academic institutions, clinical research organizations, and biotechnology companies.

In the field of cellular biology, single-cell analysis and subcellular analysis is the study of genomics, transcriptomics, proteomics, metabolomics and cell–cell interactions at the single cell level. The concept of single-cell analysis originated in the 1970s. Before the discovery of heterogeneity, single-cell analysis mainly referred to the analysis or manipulation of an individual cell in a bulk population of cells at a particular condition using optical or electronic microscope. To date, due to the heterogeneity seen in both eukaryotic and prokaryotic cell populations, analyzing a single cell makes it possible to discover mechanisms not seen when studying a bulk population of cells. Technologies such as fluorescence-activated cell sorting (FACS) allow the precise isolation of selected single cells from complex samples, while high throughput single cell partitioning technologies, enable the simultaneous molecular analysis of hundreds or thousands of single unsorted cells; this is particularly useful for the analysis of transcriptome variation in genotypically identical cells, allowing the definition of otherwise undetectable cell subtypes. The development of new technologies is increasing our ability to analyze the genome and transcriptome of single cells, as well as to quantify their proteome and metabolome. Mass spectrometry techniques have become important analytical tools for proteomic and metabolomic analysis of single cells. Recent advances have enabled quantifying thousands of protein across hundreds of single cells, and thus make possible new types of analysis. In situ sequencing and fluorescence in situ hybridization (FISH) do not require that cells be isolated and are increasingly being used for analysis of tissues.
Single-cell sequencing examines the nucleic acid sequence information from individual cells with optimized next-generation sequencing technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment. For example, in cancer, sequencing the DNA of individual cells can give information about mutations carried by small populations of cells. In development, sequencing the RNAs expressed by individual cells can give insight into the existence and behavior of different cell types. In microbial systems, a population of the same species can appear genetically clonal. Still, single-cell sequencing of RNA or epigenetic modifications can reveal cell-to-cell variability that may help populations rapidly adapt to survive in changing environments.

Standard BioTools Inc., previously known as Fluidigm Corp., offers analytical mass cytometry systems for flow cytometry and tissue imaging, along with associated assays and reagents, as well as an automated genomic analysis instrument and a variety of microfluidic arrays, or integrated fluidic circuits (IFCs), and consumables with fully kitted reagents. Custom assays and services are available with all systems and applications.
Perturb-seq refers to a high-throughput method of performing single cell RNA sequencing (scRNA-seq) on pooled genetic perturbation screens. Perturb-seq combines multiplexed CRISPR mediated gene inactivations with single cell RNA sequencing to assess comprehensive gene expression phenotypes for each perturbation. Inferring a gene’s function by applying genetic perturbations to knock down or knock out a gene and studying the resulting phenotype is known as reverse genetics. Perturb-seq is a reverse genetics approach that allows for the investigation of phenotypes at the level of the transcriptome, to elucidate gene functions in many cells, in a massively parallel fashion.
Single-cell transcriptomics examines the gene expression level of individual cells in a given population by simultaneously measuring the RNA concentration of hundreds to thousands of genes. Single-cell transcriptomics makes it possible to unravel heterogeneous cell populations, reconstruct cellular developmental pathways, and model transcriptional dynamics — all previously masked in bulk RNA sequencing.
Droplet-based microfluidics manipulate discrete volumes of fluids in immiscible phases with low Reynolds number and laminar flow regimes. Interest in droplet-based microfluidics systems has been growing substantially in past decades. Microdroplets offer the feasibility of handling miniature volumes of fluids conveniently, provide better mixing, encapsulation, sorting, sensing and are suitable for high throughput experiments. Two immiscible phases used for the droplet based systems are referred to as the continuous phase and dispersed phase.
CITE-Seq is a method for performing RNA sequencing along with gaining quantitative and qualitative information on surface proteins with available antibodies on a single cell level. So far, the method has been demonstrated to work with only a few proteins per cell. As such, it provides an additional layer of information for the same cell by combining both proteomics and transcriptomics data. For phenotyping, this method has been shown to be as accurate as flow cytometry by the groups that developed it. It is currently one of the main methods, along with REAP-Seq, to evaluate both gene expression and protein levels simultaneously in different species.
snRNA-seq, also known as single nucleus RNA sequencing, single nuclei RNA sequencing or sNuc-seq, is an RNA sequencing method for profiling gene expression in cells which are difficult to isolate, such as those from tissues that are archived or which are hard to be dissociated. It is an alternative to single cell RNA seq (scRNA-seq), as it analyzes nuclei instead of intact cells.
Aaron R. Wheeler is a Canadian chemist who is a professor of chemistry and biomedical engineering at the University of Toronto since 2005 with cross-appointment at Institute of Biomedical Engineering and Terrence Donnelly Centre for Cellular and Biomolecular Research. His academic laboratory is located at Lash Miller Chemical Laboratories and Terrence Donnelly Centre for Cellular and Biomolecular Research at the University of Toronto. In 2005, Wheeler was appointed as assistant professor and Tier II Canada Research Chair then promoted to associate professor in 2010, full professor in 2013, and in 2018 he became the Tier I Canada Research Chair in Microfluidic Bioanalysis.

Christoph Merten is a German bio-engineer and entrepreneur; currently professor at EPFL. He is an adjunct scientist at the Ludwig Institute for Cancer Research in Lausanne. His research focuses on developing biomedical microfluidics technologies for drug discovery, diagnostics, and personalized therapy in cancer research.

MicroRNA (miRNA) biosensors are analytical devices that involve interactions between the target miRNA strands and recognition element on a detection platform to produce signals that can be measured to indicate levels or the presence of the target miRNA. Research into miRNA biosensors shows shorter readout times, increased sensitivity and specificity of miRNA detection and lower fabrication costs than conventional miRNA detection methods.









