
Emma Schymanski (born Emma Craven) is chemist known for her work identifying unknown organic compounds, particularly pollutants, and is an advocate for open science.

Emma Schymanski (born Emma Craven) is chemist known for her work identifying unknown organic compounds, particularly pollutants, and is an advocate for open science.
Schymanski graduated with a B.Sc. in Chemistry and a B.E. in Environmental Engineering from the University of Western Australia in 2003. [1] While at the University of Western Australia, Schymanski combined chemistry and environmental engineering to study contaminated sites that required assessment and remediation. [1] As an undergraduate, she participated in the 2002 Nobel Laureate conference which brings Nobel laureates and young scientists together; Schymanski and Pia Sappl were the first students from the University of Western Australia to receive this invitation and possibly the first Australians. [2]
After college, Schymanski spent three years at Golder Associates in Perth as an environmental engineer [3] and then joined the Helmholtz Centre for Environmental Research in Leipzig Germany where she finished her Ph.D. in 2011. [3] Schymanski's subsequent postdoctoral position was at the Swiss Federal Institute of Aquatic Science and Technology (Eawag) including a Marie Curie Intra-European Postdoctoral Fellowship. [3] Schymanski is currently an Associate Professor at the University of Luxembourg where she is the head of the Environmental Cheminformatics Group. [4]
In 2021, Schymanski was interviewed by the Metabolomics Society article in MetaboNews and during the interview she describes her introduction to the field of metabolomics, the current strengths of the field, and potential future applications of metabolomics research. [5]
Schymanski's first research publications were from her undergraduate work when she worked on developing new metal-containing polymers which resulted in three lead author publications. [6] [7] [8]
As a graduate student, Schymanski started using information on the fragmentation pattern of organic compounds as a means to expand the identification of unknown compounds. [9] Schymanski applied these novel methods to the identification of unknown organic compounds found in wastewater, [10] and used data collectively gathered by the NORMAN Association to define barriers to the identification of unknown organic compounds in water. [11] Identifying and tracking unknown organic compounds continues to be an avenue of research pursued by Schymanski [12] and she is a co-author on a 2014 textbook describing these methods. [13]
In 2012, Schymanski and Steffen Neumann started the Critical Assessment of Small Molecule Identification (CASMI) contest that provided researchers with information about unknown organic compounds and challenged them to use automated computational tools identify the unknown compounds. [14] [15] The Metabolomics Society highlighted the 2012 contest in their newsletter. [16] There have been multiple iterations of the contest, and Schymanski examined the results of the 2016 contest. [17]
Schymanski's research focuses on characterizing organic compounds found in wastewater [10] [18] and exposomics, or the science of compounds that people are exposed to over their lifetimes. [19] Schymanski has developed a subset of PubChem for exposomics, PubChemLite, which can be annotated to increase ability of researchers to identify unknown environmental compounds. [20] Within this field, Schymanski is working to automate the identification of a group of fluorinated compounds called ‘per- and poly-fluoroalkyl substances’ (PFASs) in order to increase the ability of researchers to find unknown PFAS in the environment. [21]
Schymanski is an advocate for open science and data sharing. Within the NORMAN network, a collaborative activity across Europe, North America, and Asia, Schymanski worked in 2011 with the team that established NORMAN MassBank, [22] [23] which was a community-driven project to gather information about small molecules. In 2015, Schymanski expanded this type of data with the NORMAN Suspect List Exchange. [24] Schymanski has also worked to develop computational tools that allow the processing of complex high resolution mass spectrometry data [25] and sought to establish standards to consider the quality of the mass spectrometry data. [26] Schymanski's 2014 publication in Environmental Science & Technology [27] establishes a means to estimate confidence in the quality of unknown organic compound identifications and, as of 2021, has over 1000 citations. In 2018, Schymanski considered this paper her greatest achievement because it established the standard for compound identification in metabolomics and encouraged community conversation about future of these tools. [28]
Schymanski is married to Stan Schymanski, an ecohydrologist. [34] and he has shared insight about dual career couples and their path to positions in Luxembourg. [35] They are the first dual career couple to both receive the FNR ATTRACT award. [32]
{{cite book}}: CS1 maint: location missing publisher (link){{cite web}}: Missing or empty |title= (help)
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a mass spectrum, a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures.

Electron ionization is an ionization method in which energetic electrons interact with solid or gas phase atoms or molecules to produce ions. EI was one of the first ionization techniques developed for mass spectrometry. However, this method is still a popular ionization technique. This technique is considered a hard ionization method, since it uses highly energetic electrons to produce ions. This leads to extensive fragmentation, which can be helpful for structure determination of unknown compounds. EI is the most useful for organic compounds which have a molecular weight below 600. Also, several other thermally stable and volatile compounds in solid, liquid and gas states can be detected with the use of this technique when coupled with various separation methods.

Gas chromatography–mass spectrometry (GC–MS) is an analytical method that combines the features of gas-chromatography and mass spectrometry to identify different substances within a test sample. Applications of GC–MS include drug detection, fire investigation, environmental analysis, explosives investigation, food and flavor analysis, and identification of unknown samples, including that of material samples obtained from planet Mars during probe missions as early as the 1970s. GC–MS can also be used in airport security to detect substances in luggage or on human beings. Additionally, it can identify trace elements in materials that were previously thought to have disintegrated beyond identification. Like liquid chromatography–mass spectrometry, it allows analysis and detection even of tiny amounts of a substance.

Metabolomics is the scientific study of chemical processes involving metabolites, the small molecule substrates, intermediates, and products of cell metabolism. Specifically, metabolomics is the "systematic study of the unique chemical fingerprints that specific cellular processes leave behind", the study of their small-molecule metabolite profiles. The metabolome represents the complete set of metabolites in a biological cell, tissue, organ, or organism, which are the end products of cellular processes. Messenger RNA (mRNA), gene expression data, and proteomic analyses reveal the set of gene products being produced in the cell, data that represents one aspect of cellular function. Conversely, metabolic profiling can give an instantaneous snapshot of the physiology of that cell, and thus, metabolomics provides a direct "functional readout of the physiological state" of an organism. There are indeed quantifiable correlations between the metabolome and the other cellular ensembles, which can be used to predict metabolite abundances in biological samples from, for example mRNA abundances. One of the ultimate challenges of systems biology is to integrate metabolomics with all other -omics information to provide a better understanding of cellular biology.

The metabolome refers to the complete set of small-molecule chemicals found within a biological sample. The biological sample can be a cell, a cellular organelle, an organ, a tissue, a tissue extract, a biofluid or an entire organism. The small molecule chemicals found in a given metabolome may include both endogenous metabolites that are naturally produced by an organism as well as exogenous chemicals that are not naturally produced by an organism.

In mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy-absorbing matrix to create ions from large molecules with minimal fragmentation. It has been applied to the analysis of biomolecules and various organic molecules, which tend to be fragile and fragment when ionized by more conventional ionization methods. It is similar in character to electrospray ionization (ESI) in that both techniques are relatively soft ways of obtaining ions of large molecules in the gas phase, though MALDI typically produces far fewer multi-charged ions.

Liquid chromatography–mass spectrometry (LC–MS) is an analytical chemistry technique that combines the physical separation capabilities of liquid chromatography with the mass analysis capabilities of mass spectrometry (MS). Coupled chromatography – MS systems are popular in chemical analysis because the individual capabilities of each technique are enhanced synergistically. While liquid chromatography separates mixtures with multiple components, mass spectrometry provides spectral information that may help to identify each separated component. MS is not only sensitive, but provides selective detection, relieving the need for complete chromatographic separation. LC–MS is also appropriate for metabolomics because of its good coverage of a wide range of chemicals. This tandem technique can be used to analyze biochemical, organic, and inorganic compounds commonly found in complex samples of environmental and biological origin. Therefore, LC–MS may be applied in a wide range of sectors including biotechnology, environment monitoring, food processing, and pharmaceutical, agrochemical, and cosmetic industries. Since the early 2000s, LC–MS has also begun to be used in clinical applications.

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important method for the accurate mass determination and characterization of proteins, and a variety of methods and instrumentations have been developed for its many uses. Its applications include the identification of proteins and their post-translational modifications, the elucidation of protein complexes, their subunits and functional interactions, as well as the global measurement of proteins in proteomics. It can also be used to localize proteins to the various organelles, and determine the interactions between different proteins as well as with membrane lipids.

Ambient ionization is a form of ionization in which ions are formed in an ion source outside the mass spectrometer without sample preparation or separation. Ions can be formed by extraction into charged electrospray droplets, thermally desorbed and ionized by chemical ionization, or laser desorbed or ablated and post-ionized before they enter the mass spectrometer.
The Kendrick mass is defined by setting the mass of a chosen molecular fragment, typically CH2, to an integer value in amu (atomic mass units). It is different from the IUPAC definition, which is based on setting the mass of 12C isotope to exactly 12 amu. The Kendrick mass is often used to identify homologous compounds differing only by a number of base units in high resolution mass spectra. This definition of mass was first suggested in 1963 by chemist Edward Kendrick, and it has been adopted by scientists working in the area of high-resolution mass spectrometry, environmental analysis, proteomics, petroleomics, metabolomics, polymer analysis, etc.

In mass spectrometry, fragmentation is the dissociation of energetically unstable molecular ions formed from passing the molecules mass spectrum. These reactions are well documented over the decades and fragmentation patterns are useful to determine the molar weight and structural information of unknown molecules. Fragmentation that occurs in tandem mass spectrometry experiments has been a recent focus of research, because this data helps facilitate the identification of molecules.
The METLIN Metabolite and Chemical Entity Database is the largest repository of experimental tandem mass spectrometry and neutral loss data acquired from standards. The tandem mass spectrometry data on over 930,000 molecular standards is provided to facilitate the identification of chemical entities from tandem mass spectrometry experiments. In addition to the identification of known molecules, it is also useful for identifying unknowns using its similarity searching technology. All tandem mass spectrometry data comes from the experimental analysis of standards at multiple collision energies and in both positive and negative ionization modes.
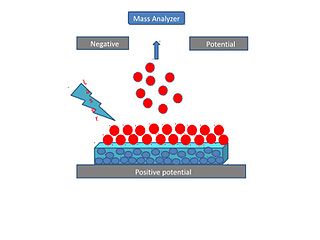
Surface-assisted laser desorption/ionization (SALDI) is a soft laser desorption technique used for mass spectrometry analysis of biomolecules, polymers, and small organic molecules. In its first embodiment Koichi Tanaka used a cobalt/glycerol liquid matrix and subsequent applications included a graphite/glycerol liquid matrix as well as a solid surface of porous silicon. The porous silicon represents the first matrix-free SALDI surface analysis allowing for facile detection of intact molecular ions, these porous silicon surfaces also facilitated the analysis of small molecules at the yoctomole level. At present laser desorption/ionization methods using other inorganic matrices such as nanomaterials are often regarded as SALDI variants. As an example, silicon nanowires as well as Titania nanotube arrays (NTA) have been used as substrates to detect small molecules. SALDI is used to detect proteins and protein-protein complexes. A related method named "ambient SALDI" - which is a combination of conventional SALDI with ambient mass spectrometry incorporating the direct analysis real time (DART) ion source has also been demonstrated. SALDI is considered one of the most important techniques in MS and has many applications.

In the field of cellular biology, single-cell analysis and subcellular analysis is the study of genomics, transcriptomics, proteomics, metabolomics and cell–cell interactions at the single cell level. The concept of single-cell analysis originated in the 1970s. Before the discovery of heterogeneity, single-cell analysis mainly referred to the analysis or manipulation of an individual cell in a bulk population of cells at a particular condition using optical or electronic microscope. To date, due to the heterogeneity seen in both eukaryotic and prokaryotic cell populations, analyzing a single cell makes it possible to discover mechanisms not seen when studying a bulk population of cells. Technologies such as fluorescence-activated cell sorting (FACS) allow the precise isolation of selected single cells from complex samples, while high throughput single cell partitioning technologies, enable the simultaneous molecular analysis of hundreds or thousands of single unsorted cells; this is particularly useful for the analysis of transcriptome variation in genotypically identical cells, allowing the definition of otherwise undetectable cell subtypes. The development of new technologies is increasing our ability to analyze the genome and transcriptome of single cells, as well as to quantify their proteome and metabolome. Mass spectrometry techniques have become important analytical tools for proteomic and metabolomic analysis of single cells. Recent advances have enabled quantifying thousands of protein across hundreds of single cells, and thus make possible new types of analysis. In situ sequencing and fluorescence in situ hybridization (FISH) do not require that cells be isolated and are increasingly being used for analysis of tissues.
Petroleomics is the identification of the totality of the constituents of naturally occurring petroleum and crude oil using high resolution mass spectrometry. In addition to mass determination, petroleomic analysis sorts the chemical compounds into heteroatom class, type. The name is a combination of petroleum and -omics.
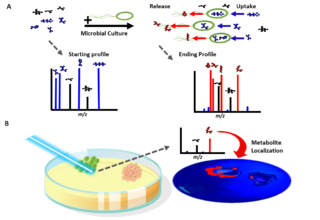
Exometabolomics, also known as 'metabolic footprinting', is the study of extracellular metabolites and is a sub-field of metabolomics.
Jennifer S. Brodbelt is an American chemist known for her research using mass spectrometry to characterize organic compounds, especially biopolymers and proteins.

The CompTox Chemicals Dashboard is a freely accessible online database created and maintained by the U.S. Environmental Protection Agency (EPA). The database provides access to multiple types of data including physicochemical properties, environmental fate and transport, exposure, usage, in vivo toxicity, and in vitro bioassay. EPA and other scientists use the data and models contained within the dashboard to help identify chemicals that require further testing and reduce the use of animals in chemical testing. The Dashboard is also used to provide public access to information from EPA Action Plans, e.g. around perfluorinated alkylated substances.

Gary Siuzdak is an American chemist best known for his work in the field of metabolomics, activity metabolomics, and mass spectrometry. His lab discovered indole-3-propionic acid as a gut bacteria derived metabolite in 2009. He is currently the Professor and Director of The Center for Metabolomics and Mass Spectrometry at Scripps Research in La Jolla, California. Siuzdak has also made contributions to virus analysis, viral structural dynamics, as well as developing mass spectrometry imaging technology using nanostructured surfaces. The Siuzdak lab is also responsible for creating the research tools eXtensible Computational Mass Spectrometry (XCMS), METLIN, METLIN Neutral Loss and Q-MRM. As of January 2021, the XCMS/METLIN platform has over 50,000 registered users.

Secondary electro-spray ionization (SESI) is an ambient ionization technique for the analysis of trace concentrations of vapors, where a nano-electrospray produces charging agents that collide with the analyte molecules directly in gas-phase. In the subsequent reaction, the charge is transferred and vapors get ionized, most molecules get protonated and deprotonated. SESI works in combination with mass spectrometry or ion-mobility spectrometry.