Related Research Articles
Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like peptidoglycan layer outside the plasma membrane, the rigid cell wall characteristic of most bacteria. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.

Glycoproteins are proteins which contain oligosaccharide chains covalently attached to amino acid side-chains. The carbohydrate is attached to the protein in a cotranslational or posttranslational modification. This process is known as glycosylation. Secreted extracellular proteins are often glycosylated.

Teichoic acids are bacterial copolymers of glycerol phosphate or ribitol phosphate and carbohydrates linked via phosphodiester bonds.
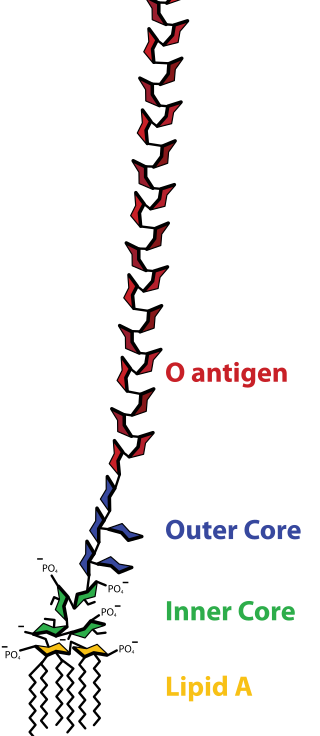
Lipopolysaccharides (LPS) are large molecules consisting of a lipid and a polysaccharide that are bacterial toxins. They are composed of an O-antigen, an outer core, and an inner core all joined by covalent bonds, and are found in the outer membrane of Gram-negative bacteria. Today, the term endotoxin is often used synonymously with LPS, although there are a few endotoxins that are not related to LPS, such as the so-called delta endotoxin proteins produced by Bacillus thuringiensis.
Dolichol refers to any of a group of long-chain mostly unsaturated organic compounds that are made up of varying numbers of isoprene units terminating in an α-saturated isoprenoid group, containing an alcohol functional group.

The bacterial outer membrane is found in gram-negative bacteria. Its composition is distinct from that of the inner cytoplasmic cell membrane - among other things, the outer leaflet of the outer membrane of many gram-negative bacteria includes a complex lipopolysaccharide whose lipid portion acts as an endotoxin - and in some bacteria such as E. coli it is linked to the cell's peptidoglycan by Braun's lipoprotein.

Glycosyltransferases are enzymes that establish natural glycosidic linkages. They catalyze the transfer of saccharide moieties from an activated nucleotide sugar to a nucleophilic glycosyl acceptor molecule, the nucleophile of which can be oxygen- carbon-, nitrogen-, or sulfur-based.

Tunicamycin is a mixture of homologous nucleoside antibiotics that inhibits the UDP-HexNAc: polyprenol-P HexNAc-1-P family of enzymes. In eukaryotes, this includes the enzyme GlcNAc phosphotransferase (GPT), which catalyzes the transfer of N-acetylglucosamine-1-phosphate from UDP-N-acetylglucosamine to dolichol phosphate in the first step of glycoprotein synthesis. Tunicamycin blocks N-linked glycosylation (N-glycans) and treatment of cultured human cells with tunicamycin causes cell cycle arrest in G1 phase. It is used as an experimental tool in biology, e.g. to induce unfolded protein response. Tunicamycin is produced by several bacteria, including Streptomyces clavuligerus and Streptomyces lysosuperificus.

The enzyme UDP-glucose 4-epimerase, also known as UDP-galactose 4-epimerase or GALE, is a homodimeric epimerase found in bacterial, fungal, plant, and mammalian cells. This enzyme performs the final step in the Leloir pathway of galactose metabolism, catalyzing the reversible conversion of UDP-galactose to UDP-glucose. GALE tightly binds nicotinamide adenine dinucleotide (NAD+), a co-factor required for catalytic activity.
Nucleotide sugars are the activated forms of monosaccharides. Nucleotide sugars act as glycosyl donors in glycosylation reactions. Those reactions are catalyzed by a group of enzymes called glycosyltransferases.
In enzymology, a lipopolysaccharide N-acetylmannosaminouronosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a N-acetyllactosaminide 3-alpha-galactosyltransferase is an enzyme that catalyzes the chemical reaction

UDP-N-acetylglucosamine—dolichyl-phosphate N-acetylglucosaminephosphotransferase is an enzyme that in humans is encoded by the DPAGT1 gene.
DTDP-4-amino-4,6-dideoxy-D-galactose acyltransferase is an enzyme with systematic name acetyl-CoA:dTDP-4-amino-4,6-dideoxy-alpha-D-galactose N-acetyltransferase. This enzyme catalyses the following chemical reaction
D-Man-alpha-(1->3) -D-Glc-beta-(1->4) -D-Glc-alpha-1-diphosphoundecaprenol 2-beta-glucuronyltransferase is an enzyme with the systematic name UDP-glucuronate: D-Man-alpha-(1->3) -D-Glc-beta-(1->4)-D-Glc-alpha-1-diphospho-ditrans,octacis-undecaprenol beta-1,2-glucuronyltransferase. This enzyme catalyses the following chemical reaction:
UDP-N-acetylglucosamine—undecaprenyl-phosphate N-acetylglucosaminephosphotransferase is an enzyme with systematic name UDP-N-acetyl-alpha-D-glucosamine:ditrans,octacis-undecaprenyl phosphate N-acetyl-alpha-D-glucosaminephosphotransferase. This enzyme catalyses the following chemical reaction

Lipid II is a precursor molecule in the synthesis of the cell wall of bacteria. It is a peptidoglycan, which is amphipathic and named for its bactoprenol hydrocarbon chain, which acts as a lipid anchor, embedding itself in the bacterial cell membrane. Lipid II must translocate across the cell membrane to deliver and incorporate its disaccharide-pentapeptide "building block" into the peptidoglycan mesh. Lipid II is the target of several antibiotics.
The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase superfamily is a group of integral membrane protein families. The MOP flippase superfamily includes twelve distantly related families, six for which functional data are available:
- One ubiquitous family (MATE) specific for drugs - (TC# 2.A.66.1) The Multi Antimicrobial Extrusion (MATE) Family
- One (PST) specific for polysaccharides and/or their lipid-linked precursors in prokaryotes - (TC# 2.A.66.2) The Polysaccharide Transport (PST) Family
- One (OLF) specific for lipid-linked oligosaccharide precursors of glycoproteins in eukaryotes - (TC# 2.A.66.3) The Oligosaccharidyl-lipid Flippase (OLF) Family
- One (MVF) lipid-peptidoglycan precursor flippase involved in cell wall biosynthesis - (TC# 2.A.66.4) The Mouse Virulence Factor (MVF) Family
- One (AgnG) which includes a single functionally characterized member that extrudes the antibiotic, Agrocin 84 - (TC# 2.A.66.5) The Agrocin 84 Antibiotic Exporter (AgnG) Family
- And finally, one (Ank) that shuttles inorganic pyrophosphate (PPi) - (TC# 2.A.66.9) The Progressive Ankylosis (Ank) Family

Methyltransferase/kinase WbdD EC 2.1.1.294 and EC 2.7.1.181WbdD is a bifunctional enzyme that regulates the length of the LPS O-antigen polysaccharide chain. Stops the polymerization of the chain by phosphorylating and then methylating the phosphate on the terminal sugar. This terminal modification is essential for export of the O-antigen across the inner membrane. WbdD is also required for correct localization of the WbdA mannosyltransferase.
Undecaprenyl phosphate (UP), also known lipid-P, bactoprenol and C55-P., is a molecule with the primary function of trafficking polysaccharides across the cell membrane, largely contributing to the overall structure of the cell wall in Gram-positive bacteria. In some situations, UP can also be utilized to carry other cell-wall polysaccharides, but UP is the designated lipid carrier for peptidoglycan. During the process of carrying the peptidoglycan across the cell membrane, N-acetylglucosamine and N-acetylmuramic acid are linked to UP on the cytoplasmic side of the membrane before being carried across. UP works in a cycle of phosphorylation and dephosphorylation as the lipid carrier gets used, recycled, and reacts with undecaprenyl phosphate. This type of synthesis is referred to as de novo synthesis where a complex molecule is created from simpler molecules as opposed to a complete recycle of the entire structure.
References
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 Rai, A. K., & Mitchell, A. M. (2020). Enterobacterial Common Antigen: Synthesis and Function of an Enigmatic Molecule. MBio, 11(4), e01914-20. https://doi.org/10.1128/mBio.01914-20
- 1 2 Kunin, C. M.; Beard, M. V.; Halmagyi, N. E. (962). "Evidence for a Common Hapten Associated with Endotoxin Fractions of E. coli and other Enterobacteriaceae". Experimental Biology and Medicine. 111 (1): 160–166. doi:10.3181/00379727-111-27734. S2CID 86972326.
- ↑ Rahman, A., Barr, K., & Rick, P. D. (2001). Identification of the Structural Gene for the TDP-Fuc4NAc:Lipid II Fuc4NAc Transferase Involved in Synthesis of Enterobacterial Common Antigen in Escherichia coli K-12. Journal of Bacteriology, 183(22), 6509–6516. https://doi.org/10.1128/JB.183.22.6509-6516.2001
- 1 2 3 Kuhn, H.-M., Meier-Dieter, U., & Mayer, H. (1988). ECA, the enterobacterial common antigen. FEMS Microbiology Letters, 54(3), 195–222.
- ↑ Castelli, M. E., & Véscovi, E. G. (2011). The Rcs Signal Transduction Pathway Is Triggered by Enterobacterial Common Antigen Structure Alterations in Serratia marcescens. Journal of Bacteriology, 193(1), 63–74. https://doi.org/10.1128/JB.00839-10
- 1 2 Shen, X., Yang, Y., Li, P., Luo, H., & Kong, Q. (2021). Advances in the Research of Enterobacterial Common Antigen. Chinese Journal of Biotechnology, 37(4), 1081–1091. https://doi.org/10.13345/j.cjb.200334
- 1 2 Kajimura, J., Rahman, A., & Rick, P. D. (2005). Assembly of Cyclic Enterobacterial Common Antigen in Escherichia coli K-12. Journal of Bacteriology, 187(20), 6917–6927. https://doi.org/10.1128/JB.187.20.6917-6927.2005