
The Wellcome Sanger Institute, previously known as The Sanger Centre and Wellcome Trust Sanger Institute, is a non-profit British genomics and genetics research institute, primarily funded by the Wellcome Trust.
The Rat Genome Database (RGD) is a database of rat genomics, genetics, physiology and functional data, as well as data for comparative genomics between rat, human and mouse. RGD is responsible for attaching biological information to the rat genome via structured vocabulary, or ontology, annotations assigned to genes and quantitative trait loci (QTL), and for consolidating rat strain data and making it available to the research community. RGD is working with groups such as the Programs for Genomic Applications at MCW and the National BioResource Project for the Rat (NBPR-Rat) in Japan to collect and make available comprehensive physiologic data for a variety of rat strains. They are also developing a suite of tools for mining and analyzing genomic, physiologic and functional data for the rat, and comparative data for rat, mouse and human.
The Zebrafish Information Network is an online biological database of information about the zebrafish. The zebrafish is a widely used model organism for genetic, genomic, and developmental studies, and ZFIN provides an integrated interface for querying and displaying the large volume of data generated by this research. To facilitate use of the zebrafish as a model of human biology, ZFIN links these data to corresponding information about other model organisms and to human disease databases. Abundant links to external sequence databases and to genome browsers are included. Gene product, gene expression, and phenotype data are annotated with terms from biomedical ontologies. ZFIN is based at the University of Oregon in the United States, with funding provided by the National Institutes of Health (NIH).

E3 ubiquitin-protein ligase RAD18 is an enzyme that in humans is encoded by the RAD18 gene.

Ubiquitin-associated protein 1 is a protein that in humans is encoded by the UBAP1 gene.

Interferon-induced transmembrane protein 3 (IFITM3) is a protein that in humans is encoded by the IFITM3 gene. It plays a critical role in the immune system's defense against Swine Flu, where heightened levels of IFITM3 keep viral levels low, and the removal of IFITM3 allows the virus to multiply unchecked. This observation has been further advanced by a recent study from Paul Kellam's lab that shows that a single nucleotide polymorphism in the human IFITM3 gene purported to increase influenza susceptibility is overrepresented in people hospitalised with pandemic H1N1. The prevalence of this mutation is thought to be approximately 1/400 in European populations.

ATP synthase mitochondrial F1 complex assembly factor 2 is an enzyme that in humans is encoded by the ATPAF2 gene.

Rho-related BTB domain-containing protein 3 is a protein that in humans is encoded by the RHOBTB3 gene.

Protein YIPF1 is a protein that in humans is encoded by the YIPF1 gene.

ATP-dependent RNA helicase SUPV3L1, mitochondrial is an enzyme that in humans is encoded by the SUPV3L1 gene.

Pseudouridylate synthase 7 homolog-like protein is an enzyme that in humans is encoded by the PUS7L gene.

SLX4 is a protein involved in DNA repair, where it has important roles in the final steps of homologous recombination. Mutations in the gene are associated with the disease Fanconi anemia.

Heterochromatin protein 1, binding protein 3 is a protein that in humans is encoded by the HP1BP3 gene. It has been identified as a novel subtype of the linker histone H1, involved in the structure of heterochromatin

Transcription factor 7-like 1 , also known as TCF7L1, is a human gene.

NOP2/Sun domain family, member 2 is a protein that in humans is encoded by the NSUN2 gene. Alternatively spliced transcript variants encoding different isoforms have been noted for the gene.
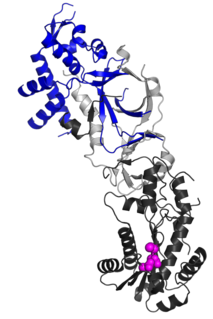
Ribonuclease H2, subunit B is a protein that in humans is encoded by the RNASEH2B gene. RNase H2 is composed of a single catalytic subunit (A) and two non-catalytic subunits, and degrades the RNA of RNA:DNA hybrids. The non-catalytic B subunit of RNase H2 is thought to play a role in DNA replication.
Uberon is a comparative anatomy ontology representing a variety of structures found in animals, such as lungs, muscles, bones, feathers and fins. These structures are connected to other structures via relationships such as part-of and develops-from. One of the uses of this ontology is to integrate data from different biological databases, and other species-specific ontologies such as the Foundational Model of Anatomy.

The International Mouse Phenotyping Consortium (IMPC) is an international scientific endeavour to create and characterize the phenotype of 20,000 knockout mouse strains. Launched in September 2011, the consortium consists of over 15 research institutes across four continents with funding provided by the NIH, European national governments and the partner institutions.
The Mouse Genetics Project (MGP) is a large-scale mutant mouse production and phenotyping programme aimed at identifying new model organisms of disease.
The Monarch Initiative is a large scale bioinformatics web resource focused on leveraging existing biomedical knowledge to connect genotypes with phenotypes in an effort to aid research that combats genetic diseases. Monarch does this by integrating multi-species genotype, phenotype, genetic variant and disease knowledge from various existing biomedical data resources into a centralized and structured database. While this integration process has been traditionally done manually by basic researchers and clinicians on a case by case basis, The Monarch Initiative provides an aggregated and structured collection of data and tools that make biomedical knowledge exploration more efficient and effective.












