An allele is one of two, or more, forms of a given gene variant. For example, the ABO blood grouping is controlled by the ABO gene, which has six common alleles. Nearly every living human's phenotype for the ABO gene is some combination of just these six alleles. An allele is one of two, or more, versions of the same gene at the same place on a chromosome. It can also refer to different sequence variations for a several-hundred base-pair or more region of the genome that codes for a protein. Alleles can come in different extremes of size. At the lowest possible size an allele can be a single nucleotide polymorphism (SNP). At the higher end, it can be up to several thousand base-pairs long. Most alleles result in little or no observable change in the function of the protein the gene codes for.
Allele frequency, or gene frequency, is the relative frequency of an allele at a particular locus in a population, expressed as a fraction or percentage. Specifically, it is the fraction of all chromosomes in the population that carry that allele. Microevolution is the change in allele frequencies that occurs over time within a population.
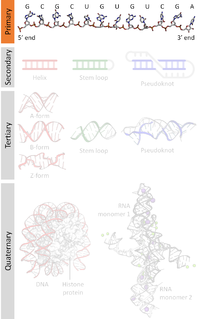
A nucleic acid sequence is a succession of bases signified by a series of a set of five different letters that indicate the order of nucleotides forming alleles within a DNA or RNA (GACU) molecule. By convention, sequences are usually presented from the 5' end to the 3' end. For DNA, the sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the primary structure.

Genetic variation is the difference in DNA among individuals or the differences between populations. There are multiple sources of genetic variation, including mutation and genetic recombination. The mutation is the ultimate source of genetic variation, but mechanisms such as sexual reproduction and genetic drift contribute to it as well.

In genetics, a single-nucleotide polymorphism is a substitution of a single nucleotide at a specific position in the genome that is present in a sufficiently large fraction of the population.
Reactome is a free online database of biological pathways. There are several Reactomes that concentrate on specific organisms, the largest of these is focused on human biology, the following description concentrates on the human Reactome. It is authored by expert biologists, in collaboration with Reactome editorial staff who are all PhD level biologists. Content is cross-referenced to many bioinformatics databases. The rationale behind Reactome is to visually represent biological pathways in full mechanistic detail, while making the source data available in a computationally accessible format.
In molecular biology, SNP array is a type of DNA microarray which is used to detect polymorphisms within a population. A single nucleotide polymorphism (SNP), a variation at a single site in DNA, is the most frequent type of variation in the genome. Around 335 million SNPs have been identified in the human genome, 15 million of which are present at frequencies of 1% or higher across different populations worldwide.
Online Mendelian Inheritance in Animals (OMIA) is an online database of genes, inherited disorders and traits in more than 135 animal species. It is modelled on, and is complementary to, Online Mendelian Inheritance in Man (OMIM). It aims to provide a publicly accessible catalogue of all animal phenes, excluding those in human and mouse, for which species specific resources are already available. Authored by Professor Frank Nicholas of the University of Sydney, with some contribution from colleagues, the database contains textual information and references as well as links to relevant PubMed and Gene records at the NCBI.
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology and evolutionary biology. It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see Glossary of evolutionary biology.

The Single Nucleotide Polymorphism Database (dbSNP) is a free public archive for genetic variation within and across different species developed and hosted by the National Center for Biotechnology Information (NCBI) in collaboration with the National Human Genome Research Institute (NHGRI). Although the name of the database implies a collection of one class of polymorphisms only, it in fact contains a range of molecular variation: (1) SNPs, (2) short deletion and insertion polymorphisms (indels/DIPs), (3) microsatellite markers or short tandem repeats (STRs), (4) multinucleotide polymorphisms (MNPs), (5) heterozygous sequences, and (6) named variants. The dbSNP accepts apparently neutral polymorphisms, polymorphisms corresponding to known phenotypes, and regions of no variation. It was created in September 1998 to supplement GenBank, NCBI’s collection of publicly available nucleic acid and protein sequences.
This microRNA database and microRNA targets databases is a compilation of databases and web portals and servers used for microRNAs and their targets. MicroRNAs (miRNAs) represent an important class of small non-coding RNAs (ncRNAs) that regulate gene expression by targeting messenger RNAs.
In molecular biology, REBASE is a database of information about restriction enzymes and DNA methyltransferases. REBASE contains an extensive set of references, sites of recognition and cleavage, sequences and structures. It also contains information on the commercial availability of each enzyme. REBASE is one of the longest running biological databases having its roots in a collection of restriction enzymes maintained by Richard J. Roberts since before 1980. Since that time there have been regular descriptions of the resource in the journal Nucleic Acids Research.
The allele frequency net database is a database containing the allele frequencies of immune genes and their corresponding alleles in different populations.
The variations and drugs database (VnD) contains a comprehensive information on diseases, related genes and genetic variations, protein structures and drug information
The term microattribution is defined as "giving database accessions the same citation conventions and indices that journal articles currently enjoy". In the sense that the purpose of precise attribution is to extend the scholarly convention of giving citation credit, the provenance of a piece of scholarship is recognized to give credit and priority to a preceding author. Microattribution is thus defined as "a scholarly contribution smaller than a journal article being ascribed to a particular author" or a small scholarly contribution being ascribed to a particular author. Of course, since data accessions can describe contributions that can vastly exceed research articles in size and quality, quantum attribution or precise citation might be better terms. Barend Mons and Jan Velterop proposed nanopublications for single, attributable and machine-readable assertions in scientific literature.
The Centre for Arab Genomic Studies (CAGS) oversees genetic analyses on the populations of the Arab world. Based in Dubai, United Arab Emirates, it indicates that Arab countries have among the highest rates of genetic disorders in the world. Some 906 pathologies are endemic to the Arab states, including thalassaemia, Tourette's syndrome, Wilson's disease, Charcot-Marie-Tooth disease, mitochondrial encephalomyopathies and Niemann-Pick disease.
In bioinformatics, a Gene Disease Database is a systematized collection of data, typically structured to model aspects of reality, in a way to comprehend the underlying mechanisms of complex diseases, by understanding multiple composite interactions between phenotype-genotype relationships and gene-disease mechanisms. Gene Disease Databases integrate human gene-disease associations from various expert curated databases and text mining derived associations including Mendelian, complex and environmental diseases.
The invertebrate mitochondrial code is a genetic code used by the mitochondrial genome of invertebrates.

BacDive is a bacterial metadatabase that provides strain-linked information about bacterial and archaeal biodiversity.
The GWAS catalog is a free online database that compiles data of genome-wide association studies (GWAS), summarizing unstructured data from different literature sources into accessible high quality data. It was created by the National Human Genome Research Institute (NHGRI) in 2008 and have become a collaborative project between the NHGRI and the European Bioinformatics Institute (EBI) since 2010. As of September 2018, it has included 71,673 SNP–trait associations in 3,567 publications.




