Related Research Articles

In computer science, a data structure is a data organization and storage format that is usually chosen for efficient access to data. More precisely, a data structure is a collection of data values, the relationships among them, and the functions or operations that can be applied to the data, i.e., it is an algebraic structure about data.

In computing, a hash table is a data structure that implements an associative array, also called a dictionary or simply map; an associative array is an abstract data type that maps keys to values. A hash table uses a hash function to compute an index, also called a hash code, into an array of buckets or slots, from which the desired value can be found. During lookup, the key is hashed and the resulting hash indicates where the corresponding value is stored. A map implemented by a hash table is called a hash map.
In computer programming, a string is traditionally a sequence of characters, either as a literal constant or as some kind of variable. The latter may allow its elements to be mutated and the length changed, or it may be fixed. A string is generally considered as a data type and is often implemented as an array data structure of bytes that stores a sequence of elements, typically characters, using some character encoding. String may also denote more general arrays or other sequence data types and structures.
A string-searching algorithm, sometimes called string-matching algorithm, is an algorithm that searches a body of text for portions that match by pattern.
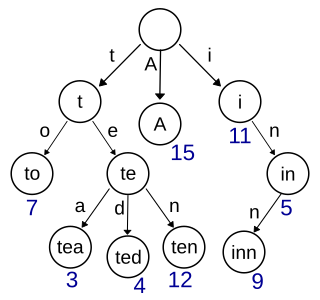
In computer science, a trie, also known as a digital tree or prefix tree, is a specialized search tree data structure used to store and retrieve strings from a dictionary or set. Unlike a binary search tree, nodes in a trie do not store their associated key. Instead, each node's position within the trie determines its associated key, with the connections between nodes defined by individual characters rather than the entire key.
In computer science, the Knuth–Morris–Pratt algorithm is a string-searching algorithm that searches for occurrences of a "word" W within a main "text string" S by employing the observation that when a mismatch occurs, the word itself embodies sufficient information to determine where the next match could begin, thus bypassing re-examination of previously matched characters.

In computer science, a suffix tree is a compressed trie containing all the suffixes of the given text as their keys and positions in the text as their values. Suffix trees allow particularly fast implementations of many important string operations.
In computer science, a suffix array is a sorted array of all suffixes of a string. It is a data structure used in, among others, full-text indices, data-compression algorithms, and the field of bibliometrics.

In computer science, a dynamic array, growable array, resizable array, dynamic table, mutable array, or array list is a random access, variable-size list data structure that allows elements to be added or removed. It is supplied with standard libraries in many modern mainstream programming languages. Dynamic arrays overcome a limit of static arrays, which have a fixed capacity that needs to be specified at allocation.
In computer science, a longest common substring of two or more strings is a longest string that is a substring of all of them. There may be more than one longest common substring. Applications include data deduplication and plagiarism detection.

In formal language theory and computer science, a substring is a contiguous sequence of characters within a string. For instance, "the best of" is a substring of "It was the best of times". In contrast, "Itwastimes" is a subsequence of "It was the best of times", but not a substring.
In computer science, a substring index is a data structure which gives substring search in a text or text collection in sublinear time. Once constructed from a document or set of documents, a substring index can be used to locate all occurrences of a pattern in time linear or near-linear in the pattern size, with no dependence or only logarithmic dependence on the document size.

In computer science, approximate string matching is the technique of finding strings that match a pattern approximately. The problem of approximate string matching is typically divided into two sub-problems: finding approximate substring matches inside a given string and finding dictionary strings that match the pattern approximately.
In computer science, empirical algorithmics is the practice of using empirical methods to study the behavior of algorithms. The practice combines algorithm development and experimentation: algorithms are not just designed, but also implemented and tested in a variety of situations. In this process, an initial design of an algorithm is analyzed so that the algorithm may be developed in a stepwise manner.
In computer science, a compressed suffix array is a compressed data structure for pattern matching. Compressed suffix arrays are a general class of data structure that improve on the suffix array. These data structures enable quick search for an arbitrary string with a comparatively small index.
In computer science, the longest common prefix array is an auxiliary data structure to the suffix array. It stores the lengths of the longest common prefixes (LCPs) between all pairs of consecutive suffixes in a sorted suffix array.

Gad Menahem Landau is an Israeli computer scientist noted for his contributions to combinatorial pattern matching and string algorithms and is the founding department chair of the Computer Science Department at the University of Haifa.
In computer science, the two-way string-matching algorithm is a string-searching algorithm, discovered by Maxime Crochemore and Dominique Perrin in 1991. It takes a pattern of size m, called a “needle”, preprocesses it in linear time O(m), producing information that can then be used to search for the needle in any “haystack” string, taking only linear time O(n) with n being the haystack's length.
In computer science, a generalized suffix array (GSA) is a suffix array containing all suffixes for a set of strings. Given the set of strings of total length , it is a lexicographically sorted array of all suffixes of each string in . It is primarily used in bioinformatics and string processing.
References
- ↑ Allauzen C., Crochemore M., Raffinot M., Factor oracle: a new structure for pattern matching; Proceedings of SOFSEM’99; Theory and Practice of Informatics.
- ↑ Assayag G., Dubnov S., Using Factor Oracles for Machine Improvisation. Soft Computing - A Fusion of Foundations, Methodologies and Applications. 2004-09-01. Springer Berlin / Heidelberg