Related Research Articles

Heredity, also called inheritance or biological inheritance, is the passing on of traits from parents to their offspring; either through asexual reproduction or sexual reproduction, the offspring cells or organisms acquire the genetic information of their parents. Through heredity, variations between individuals can accumulate and cause species to evolve by natural selection. The study of heredity in biology is genetics.
Penetrance in genetics is the proportion of individuals carrying a particular variant of a gene that also express an associated trait. In medical genetics, the penetrance of a disease-causing mutation is the proportion of individuals with the mutation who exhibit clinical symptoms among all individuals with such mutation. For example, if a mutation in the gene responsible for a particular autosomal dominant disorder has 95% penetrance, then 95% of those with the mutation will develop the disease, while 5% will not.
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be more linked than markers that are far apart. In other words, the nearer two genes are on a chromosome, the lower the chance of recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly unlinked.
A quantitative trait locus (QTL) is a locus that correlates with variation of a quantitative trait in the phenotype of a population of organisms. QTLs are mapped by identifying which molecular markers correlate with an observed trait. This is often an early step in identifying and sequencing the actual genes that cause the trait variation.
Forward genetics is a molecular genetics approach of determining the genetic basis responsible for a phenotype. Forward genetics methods begin with the identification of a phenotype, and finds or creates model organisms that display the characteristic being studied.

Y linkage, also known as holandric inheritance, describes traits that are produced by genes located on the Y chromosome. It is a form of sex linkage.
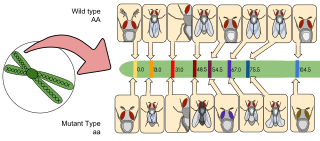
Gene mapping describes the methods used to identify the locus of a gene and the distances between genes. Gene mapping can also describe the distances between different sites within a gene.
Genetic association is when one or more genotypes within a population co-occur with a phenotypic trait more often than would be expected by chance occurrence.

In genetics, a locus is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000.

The heritability of autism is the proportion of differences in expression of autism that can be explained by genetic variation; if the heritability of a condition is high, then the condition is considered to be primarily genetic. Autism has a strong genetic basis, although the genetics of autism are complex and it is unclear whether autism spectrum disorder (ASD) is explained more by multigene interactions or by rare mutations with major effects.
Marker assisted selection or marker aided selection (MAS) is an indirect selection process where a trait of interest is selected based on a marker linked to a trait of interest, rather than on the trait itself. This process has been extensively researched and proposed for plant and animal breeding.

In genomics, a genome-wide association study, also known as whole genome association study, is an observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms.
Genetic epidemiology is the study of the role of genetic factors in determining health and disease in families and in populations, and the interplay of such genetic factors with environmental factors. Genetic epidemiology seeks to derive a statistical and quantitative analysis of how genetics work in large groups.

Like many other medical conditions, obesity is the result of an interplay between environmental and genetic factors. Studies have identified variants in several genes that may contribute to weight gain and body fat distribution; although, only in a few cases are genes the primary cause of obesity.
In genetics, association mapping, also known as "linkage disequilibrium mapping", is a method of mapping quantitative trait loci (QTLs) that takes advantage of historic linkage disequilibrium to link phenotypes to genotypes, uncovering genetic associations.
Locus heterogeneity occurs when mutations at multiple genomic loci are capable of producing the same phenotype, and each individual mutation is sufficient to cause the specific phenotype independently. Locus heterogeneity should not be confused with allelic heterogeneity, in which a single phenotype can be produced by multiple mutations, all of which are at the same locus on a chromosome. Likewise, it should not be confused with phenotypic heterogeneity, in which different phenotypes arise among organisms with identical genotypes and environmental conditions. Locus heterogeneity and allelic heterogeneity are the two components of genetic heterogeneity.
A human disease modifier gene is a modifier gene that alters expression of a human gene at another locus that in turn causes a genetic disease. Whereas medical genetics has tended to distinguish between monogenic traits, governed by simple, Mendelian inheritance, and quantitative traits, with cumulative, multifactorial causes, increasing evidence suggests that human diseases exist on a continuous spectrum between the two.
John N. Constantino is a child psychiatrist and expert on neurodevelopmental disorders, especially autism spectrum disorders (ASD). Constantino is the Blanche F. Ittleson Professor of Psychiatry and Pediatrics at Washington University School of Medicine.

Joan Ellen Bailey-Wilson is an American statistical geneticist. She is a senior investigator and co-chief of the Computational and Statistical Genomic Branch of the National Human Genome Research Institute.

Mahmud Muhieddine Barmada Known as Michael or Mike Barmada, was an American geneticist, professor, and academic administrator of Syrian-Lebanese descent. Growing his scientific career at the University of Pittsburgh apace with his field, at the beginning of high throughput and data-intensive computing, using the cloud as a research tool long before we used it to store selfies, garnering grant funding at a steady rate, mentoring over 30 masters, doctorate, medical students and faculty, Mike taught genetics, bioinformatics, and analysis at the graduate and postgraduate level and adjunct institutions. He served on over 25 committees since 2001 and as Director, Bioinformatics Resource Center, Institute for Personalized Medicine, since 2012, Associate Director, Center for Simulation and Modeling since 2012, Director, Center for Computational Genetics, GSPH since 2004, Director, Department Computational Resources Division, Department of Human Genetics, GSPH since 1999. It was his greatest joy and honor to be a teacher. He took that show on the road with Dan Weeks via the India-US Research Training Program in Genetics funded by NIH Fogarty International Center. Mike was PSIAA certified ski instructor teaching at Hidden Valley throughout the 1990s and a BSA scoutmaster for some years with Troop 834. In every interaction, Mike cherished the minds of his family, friends, collaborators, coworkers, fellow volunteers, and students.
References
- ↑ Roy, Alec; Rylander, Gunnar; Sarchiapone, Marco (December 1997). "Genetics of Suicide.: Family Studies and Molecular Genetics". Annals of the New York Academy of Sciences. 836 (1 Neurobiology): 135–157. doi:10.1111/j.1749-6632.1997.tb52358.x. ISSN 0077-8923. PMID 9616797. S2CID 29415906.
- ↑ Hopper, John L.; Bishop, D. Timothy; Easton, Douglas F. (October 2005). "Population-based family studies in genetic epidemiology". Lancet. 366 (9494): 1397–1406. doi:10.1016/S0140-6736(05)67570-8. ISSN 1474-547X. PMID 16226618. S2CID 6434807.
- ↑ Nestadt, Gerald; Samuels, Jack; Riddle, Mark; Bienvenu, O. Joseph; Liang, Kung-Yee; LaBuda, Michele; Walkup, John; Grados, Marco; Hoehn-Saric, Rudolf (2000-04-01). "A Family Study of Obsessive-compulsive Disorder". Archives of General Psychiatry. 57 (4): 358–63. doi: 10.1001/archpsyc.57.4.358 . ISSN 0003-990X. PMID 10768697.
- ↑ "familial aggregation". TheFreeDictionary.com. Retrieved 2021-11-05.
- ↑ Matthews, Abigail G.; Finkelstein, Dianne M.; Betensky, Rebecca A. (2008-10-30). "Analysis of familial aggregation studies with complex ascertainment schemes". Statistics in Medicine. 27 (24): 5076–5092. doi:10.1002/sim.3327. ISSN 0277-6715. PMC 2562890 . PMID 18618413.
- ↑ "linkage | Learn Science at Scitable". www.nature.com. Retrieved 2021-11-05.
- ↑ "3.10: Genetic Linkage". Biology LibreTexts. 2016-09-21. Retrieved 2021-11-05.
- ↑ "Family Studies in Genetics". Encyclopedia of Epidemiology. Thousand Oaks: SAGE Publications, Inc. 2008. doi:10.4135/9781412953948.n153. ISBN 9781412928168.
- ↑ "Locus". Genome.gov. Retrieved 2021-11-05.
- ↑ Ezkurdia, Iakes; Juan, David; Rodriguez, Jose Manuel; Frankish, Adam; Diekhans, Mark; Harrow, Jennifer; Vazquez, Jesus; Valencia, Alfonso; Tress, Michael L. (2014-11-15). "Multiple evidence strands suggest that there may be as few as 19 000 human protein-coding genes". Human Molecular Genetics. 23 (22): 5866–5878. doi:10.1093/hmg/ddu309. ISSN 0964-6906. PMC 4204768 . PMID 24939910.