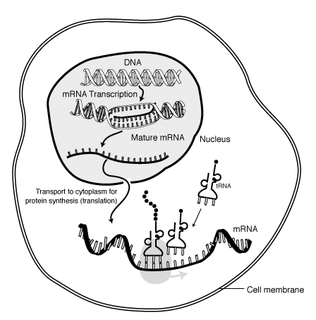
Messenger RNA (mRNA) is a large family of RNA molecules that convey genetic information from DNA to the ribosome, where they specify the amino acid sequence of the protein products of gene expression. RNA polymerase transcribes primary transcript mRNA into processed, mature mRNA. This mature mRNA is then translated into a polymer of amino acids: a protein, as summarized in the central dogma of molecular biology.
Gene knockdown is an experimental technique by which the expression of one or more of an organism's genes are reduced. The reduction can occur either through genetic modification or by treatment with a reagent such as a short DNA or RNA oligonucleotide that has a sequence complementary to either gene or an mRNA transcript.

Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein. asRNAs have been found in both prokaryotes and eukaryotes, antisense transcripts can be classified into short and long non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications.
In molecular biology ctRNA is a plasmid encoded noncoding RNA that binds to the mRNA of repB and causes translational inhibition. ctRNA is encoded by plasmids and functions in rolling circle replication to maintain a low copy number. In Corynebacterium glutamicum, it achieves this by antisense pairing with the mRNA of RepB, a replication initiation protein. In Enterococcus faecium the plasmid pJB01 contains three open reading frames, copA, repB, and repC. The pJB01 ctRNA is coded on the opposite strand from the copA/repB intergenic region and partially overlaps an atypical ribosome binding site for repB.

In molecular biology, the iron response element or iron-responsive element (IRE) is a short conserved stem-loop which is bound by iron response proteins. The IRE is found in UTRs of various mRNAs whose products are involved in iron metabolism. For example, the mRNA of ferritin contains one IRE in its 5' UTR. When iron concentration is low, IRPs bind the IRE in the ferritin mRNA and cause reduced translation rates. In contrast, binding to multiple IREs in the 3' UTR of the transferrin receptor leads to increased mRNA stability.

The micF RNA is a non-coding RNA stress response gene found in Escherichia coli and related bacteria that post-transcriptionally controls expression of the outer membrane porin gene ompF. The micF gene encodes a non-translated 93 nucleotide antisense RNA that binds its target ompF mRNA and regulates ompF expression by inhibiting translation and inducing degradation of the message. In addition, other factors, such as the RNA chaperone protein StpA also play a role in this regulatory system. Expression of micF is controlled by both environmental and internal stress factors. Four transcriptional regulators are known to bind the micF promoter region and activate micF expression.
Sib RNA refers to a group of related non-coding RNA. They were originally named QUAD RNA after they were discovered as four repeat elements in Escherichia coli intergenic regions. The family was later renamed Sib when it was discovered that the number of repeats is variable in other species and in other E. coli strains.

R1162-like plasmid antisense RNA is a 75-base RNA molecule which negatively regulates the RepI region of the plasmid. The protein product of this gene region, along with another protein, controls the copy number of the 8.75kB R1162 plasmid.
RNAI is a non-coding RNA that is an antisense repressor of the replication of some E. coli plasmids, including ColE1. Plasmid replication is usually initiated by RNAII, which acts as a primer by binding to its template DNA. The complementary RNAI binds RNAII prohibiting it from its initiation role. The rate of degradation of RNAI is therefore a major factor in the control of plasmid replication. This rate of degradation is aided by the pcnB gene product, which polyadenylates the 3' end of RNAI targeting it for degradation by PNPase.
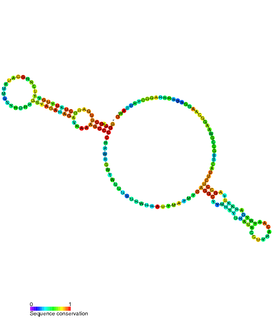
The hok/sok system is a postsegregational killing mechanism employed by the R1 plasmid in Escherichia coli. It was the first type I toxin-antitoxin pair to be identified through characterisation of a plasmid-stabilising locus. It is a type I system because the toxin is neutralised by a complementary RNA, rather than a partnered protein.

The Chlorobi-RRM RNA motif is a conserved RNA structure identified by bioinformatics. It is found within bacteria in the phylum Chlorobi, and is exclusively detected in the presumed 5' untranslated regions of genes that encode putative RNA-binding proteins. Since many RNA-binding proteins regulate their own expression in a feedback mechanism by binding or acting up their 5' UTR, it was proposed that the Chlorobi-RRM is a component in an analogous feedback mechanism. Structurally, the motif consists of two stem-loops, the second of which might function as a rho-independent transcription terminator.

The traJ-II RNA motif is a conserved RNA structure discovered in bacteria by using bioinformatics. traJ-II RNAs appear to be in the 5' untranslated regions of protein-coding genes called traJ, which functions in the process of bacterial conjugation. A previously identified motif known as TraJ 5' UTR is also found upstream of traJ genes functions as the target of FinP antisense RNAs, so it is possible that traJ-II RNAs play a similar role as targets of an antisense RNA. However, some sequence features within the traJ-II RNA motif suggest that the biological RNA might be transcribed from the reverse-complement strand. Thus is it unclear whether traJ-II function as cis-regulatory elements. traJ-II RNAs are found in a variety of proteobacteria.
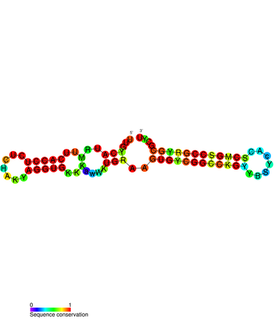
PtaRNA1 is a family of non-coding RNAs. Homologs of PtaRNA1 can be found in the proteobacteria families, Betaproteobacteria and Gammaproteobacteria. In all cases the PtaRNA1 is located anti-sense to a short protein-coding gene. In Xanthomonas campestris pv. vesicatoria, this gene is annotated as XCV2162 and is included in the plasmid toxin family of proteins.

A toxin-antitoxin system is a set of two or more closely linked genes that together encode both a "toxin" protein and a corresponding "antitoxin". When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK). Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies.
Plasmid-mediated resistance is the transfer of antibiotic resistance genes which are carried on plasmids. The plasmids can be transferred between bacteria within the same species or between different species via conjugation. Plasmids often carry multiple antibiotic resistance genes, contributing to the spread of multidrug-resistance (MDR). Antibiotic resistance mediated by MDR plasmids severely limits the treatment options for the infections caused by Gram-negative bacteria, especially Enterobacteriaceae family. The global spread of MDR plasmids has been enhanced by selective pressure from antibiotic usage in human and veterinary medicine.
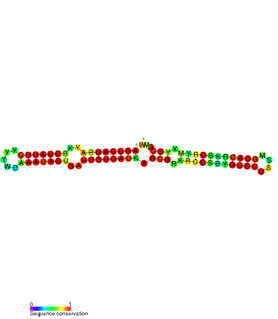
The SymE-SymR toxin-antitoxin system consists of a small hydrophobic toxin named SymE and a non-coding RNA called SymR which inhibits its translation. It is a type I toxin-antitoxin system, and is under regulation by the antitoxin, SymR.

The par stability determinant is a 400 bp locus of the pAD1 plasmid which encodes a type I toxin-antitoxin system in Enterococcus faecalis. It was the first such plasmid addiction module to be found in gram-positive bacteria.
ZEB2-AS1 is a long non-coding RNA, which is overlapping and antisense to the ZEB2 gene. It overlaps the 5' splice site of an intron within the 5'UTR of the ZEB2 gene. This intron contains an internal ribosome entry site (IRES), which is necessary for ZEB2 expression. ZEB2-AS1 prevents the splicing of this intron, and therefore activates ZEB2 expression.


















