
Hepadnaviridae is a family of viruses. Humans, apes, and birds serve as natural hosts. There are currently 18 species in this family, divided among 5 genera. Its best-known member is hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas, and cirrhosis. It is the sole accepted family in the order Blubervirales.
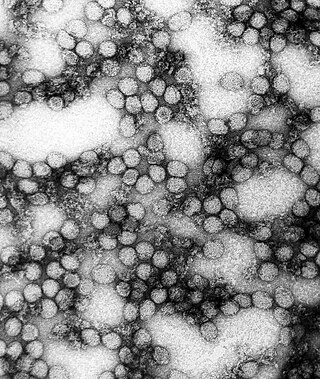
Flavivirus is a genus of positive-strand RNA viruses in the family Flaviviridae. The genus includes the West Nile virus, dengue virus, tick-borne encephalitis virus, yellow fever virus, Zika virus and several other viruses which may cause encephalitis, as well as insect-specific flaviviruses (ISFs) such as cell fusing agent virus (CFAV), Palm Creek virus (PCV), and Parramatta River virus (PaRV). While dual-host flaviviruses can infect vertebrates as well as arthropods, insect-specific flaviviruses are restricted to their competent arthropods. The means by which flaviviruses establish persistent infection in their competent vectors and cause disease in humans depends upon several virus-host interactions, including the intricate interplay between flavivirus-encoded immune antagonists and the host antiviral innate immune effector molecules.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.

Tick-borne encephalitis virus (TBEV) is a positive-strand RNA virus associated with tick-borne encephalitis in the genus Flavivirus.

RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.

Deformed wing virus (DWV) is an RNA virus, one of 22 known viruses affecting honey bees. While most commonly infecting the honey bee, Apis mellifera, it has also been documented in other bee species, like Bombus terrestris, thus, indicating it may have a wider host specificity than previously anticipated. The virus was first isolated from a sample of symptomatic honeybees from Japan in the early 1980s and is currently distributed worldwide. It is found also in pollen baskets and commercially reared bumblebees. Its main vector in A. mellifera is the Varroa mite. It is named after what is usually the most obvious deformity it induces in the development of a honeybee pupa, which is shrunken and deformed wings, but other developmental deformities are often present.
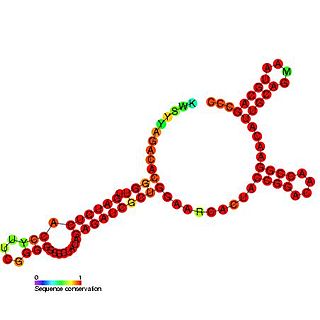
Tombusvirus 3′ UTR is an important cis-regulatory region of the Tombus virus genome.
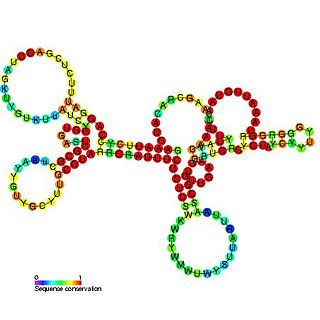
Tombusvirus 5′ UTR is an important cis-regulatory region of the Tombus virus genome.
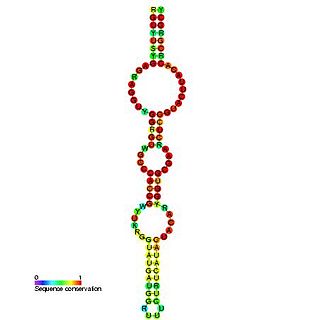
In virology, the tombusvirus internal replication element (IRE) is a segment of RNA located within the region coding for p92 polymerase. This element is essential for viral replication; specifically, it is thought to be required at an early stage of replication, such as template recruitment and/or replicase complex assembly.

The Flavivirus capsid hairpin cHP is a conserved RNA hairpin structure identified within the capsid coding region of several flavivirus genomes. These positive strand RNA genomes are translated as a single polypeptide and subsequently cleaved into constituent proteins, the first of which is the capsid protein. The cHP hairpin is located within the capsid coding region between two AUG start codons. The cHP cis element has been shown to direct translation start from the suboptimal first start codon. The ability of cHP to direct initiation from the first start codon is proportional to its thermodynamic stability, is position dependent, and is sequence independent. It has been demonstrated that both AUGs and the conserved cHP are necessary for efficient viral replication in human and mosquito cells.

Nonstructural protein 5A (NS5A) is a zinc-binding and proline-rich hydrophilic phosphoprotein that plays a key role in Hepatitis C virus RNA replication. It appears to be a dimeric form without trans-membrane helices.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3′ and 5′ untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5′ cap is attached onto mRNA and this 5′ cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3′ UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.
Entebbe bat virus is an infectious disease caused by a Flavivirus that is closely related to yellow fever.
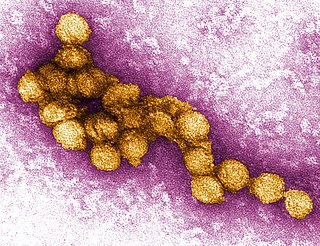
West Nile virus (WNV) is a single-stranded RNA virus that causes West Nile fever. It is a member of the family Flaviviridae, from the genus Flavivirus, which also contains the Zika virus, dengue virus, and yellow fever virus. The virus is primarily transmitted by mosquitoes, mostly species of Culex. The primary hosts of WNV are birds, so that the virus remains within a "bird–mosquito–bird" transmission cycle. The virus is genetically related to the Japanese encephalitis family of viruses.

Positive-strand RNA viruses are a group of related viruses that have positive-sense, single-stranded genomes made of ribonucleic acid. The positive-sense genome can act as messenger RNA (mRNA) and can be directly translated into viral proteins by the host cell's ribosomes. Positive-strand RNA viruses encode an RNA-dependent RNA polymerase (RdRp) which is used during replication of the genome to synthesize a negative-sense antigenome that is then used as a template to create a new positive-sense viral genome.
Yokose virus (YOKV) is in the genus Flavivirus of the family Flaviviridae. Flaviviridae are often found in arthropods, such as mosquitoes and ticks, and may also infect humans. The genus Flavivirus includes over 50 known viruses, including Yellow Fever, West Nile Virus, Zika Virus, and Japanese Encephalitis. Yokose virus is a new member of the Flavivirus family that has only been identified in a few bat species. Bats have been associated with several emerging zoonotic diseases such as Ebola and SARS.
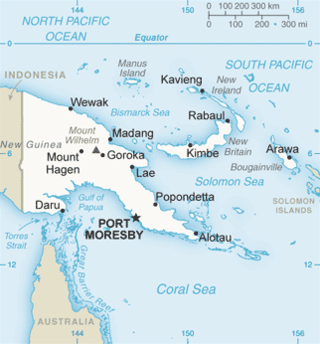
Sepik virus (SEPV) is an arthropod-borne virus (arbovirus) of the genus Flavivirus and family Flaviviridae. Flaviviridae is one of the most well characterized viral families, as it contains many well-known viruses that cause diseases that have become very prevalent in the world, like Dengue virus. The genus Flavivirus is one of the largest viral genera and encompasses over 50 viral species, including tick and mosquito borne viruses like Yellow fever virus and West Nile virus. Sepik virus is much less well known and has not been as well-classified as other viruses because it has not been known of for very long. Sepik virus was first isolated in 1966 from the mosquito Mansoniaseptempunctata, and it derives its name from the Sepik River area in Papua New Guinea, where it was first found. The geographic range of Sepik virus is limited to Papua New Guinea, due to its isolation.

Modoc virus (MODV) is a rodent-associated flavivirus. Small and enveloped, MODV contains positive single-stranded RNA. Taxonomically, MODV is part of the Flavivirus genus and Flaviviridae family. The Flavivirus genus includes nearly 80 viruses, both vector-borne and no known vector (NKV) species. Known flavivirus vector-borne viruses include Dengue virus, Yellow Fever virus, tick-borne encephalitis virus, and West Nile virus.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 5′ end which is called the 5′ UTR. The 5′ UTR is responsible for important biological functions, such as viral replication, transcription and packaging. The 5′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.
Flavivirus 3' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.
















