
PhotoRC RNA motifs refer to conserved RNA structures that are associated with genes acting in the photosynthetic reaction centre of photosynthetic bacteria. Two such RNA classes were identified and called the PhotoRC-I and PhotoRC-II motifs. PhotoRC-I RNAs were detected in the genomes of some cyanobacteria. Although no PhotoRC-II RNA has been detected in cyanobacteria, one is found in the genome of a purified phage that infects cyanobacteria. Both PhotoRC-I and PhotoRC-II RNAs are present in sequences derived from DNA that was extracted from uncultivated marine bacteria.

The D12-methyl RNA motif is a conserved RNA structure that was discovered by bioinformatics. D12-methyl motifs are found in metagenomic DNA samples, and have not yet been found in a classified organism.

The DUF2693 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2693 motif RNAs are found in Porphyromonas.

The DUF2800 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2800 motif RNAs are found in Bacillota. DUF2800 RNAs are also predicted in the phyla Actinomycetota and Synergistota, although these RNAs are likely the result of recent horizontal gene transfer or conceivably sequence contamination.

The DUF2815 RNA motif is a conserved RNA structure that was discovered by bioinformatics. As of 2018, the DUF2815 motif has not been identified in any classified organism, but is known through metagenomic sequences isolated from environmental sources.
The DUF3085 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF3085 motifs are found in one species in the genus Thauera, as well as various metagenomic sequences obtained from environmental DNA.

The DUF3577 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF3577 motifs are found in the organism Cardiobacterium valvarum and metagenomic sequences from unknown organisms.

The DUF805 RNA motif is a conserved RNA structure that was discovered by bioinformatics. The motif is subdivided into the DUF805 motif and the DUF805b motif, which have similar, but distinct secondary structures. Together, these motifs are found in Bacteroidota, Chlorobiota, and Pseudomonadota.

The GA-cis RNA motif is a conserved RNA structure that was discovered by bioinformatics. GA-cis motif RNAs are found in one species classified within the phylum Bacillota: specifically, there are 9 predicted copies in Coprocuccus eutactus ATCC 27759.

The gntR-DTE RNA motif is a conserved RNA structure that was discovered by bioinformatics. gntR-DTE motifs are found in some, but not all species within the genus Streptomyces.

The GP20 RNA motif is a conserved RNA structure that was discovered by bioinformatics. THE GP20 motif is subdivided into two related motifs, called GP20-a and GP20-b. GP20-a RNAs are found exclusively in metagenomic sequences isolated from the gut, while GP20-b RNAs are found in bacteria classified as Clostridia. Both motifs share similar, but not identical secondary structures.
lysM RNA motifs are conserved RNA structures that were discovered by bioinformatics. Such bacterial motifs are defined by consistently being upstream of 'lysM' genes, which encode lysin protein domains, a conserved domain that participates in cell wall degradation. lysM motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes, although this hypothesis is not certain.
The mcrA RNA motif is a conserved RNA structure that was discovered by bioinformatics.

The PGK RNA motif is a conserved RNA structure that was discovered by bioinformatics. PGK motif RNAs are found in metagenomic sequences isolated from the gastrointestinal tract of mammals. PGK RNAs have not yet been detected in a classified organism.
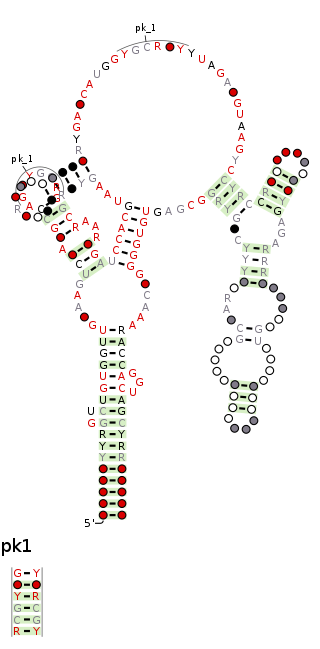
The raiA RNA motif is a conserved RNA structure that was discovered by bioinformatics. raiA motif RNAs are found in Actinomycetota and Bacillota, and have many conserved features—including conserved nucleotide positions, conserved secondary structures and associated protein-coding genes—in both of these phyla. Some conserved features of the raiA RNA motif suggest that they function as cis-regulatory elements, but other aspects of the motif suggest otherwise.
The sbcC RNA motif is a conserved RNA structure that was discovered by bioinformatics. The sbcC motif has, as of 2018, only been detected in metagenomic sequences, and the identities of organisms that contain these RNAs is unknown.

The ssNA-helicase RNA motif is a conserved RNA structure that was discovered by bioinformatics. Although the ssNA-helicase motif was published as an RNA candidate, there is some reason to suspect that it might function as a single-stranded DNA. In terms of secondary structure, RNA and DNA are difficult to distinguish when only sequence information is available.
A Streptomyces-metKH RNA motif is a conserved RNA structure that was discovered by bioinformatics. Such motifs are found in the genus Streptomyces, and are present upstream of either metK genes, which encode the S-adenosylmethionine synthetase enzyme or metH genes, which encodes the adenosylcobalamin-dependent form of methionine synthase. The RNA structures upstream of metK and metH genes are distinct from each other, but exhibit overall similar sequence and secondary structure features, suggesting that they are related to one another. Their presence upstream of protein-coding genes, and the fact that the genes perform related steps in metabolism, suggests that the RNAs function as cis-regulatory elements.

The uup RNA motif is a conserved RNA structure that was discovered by bioinformatics. uup motif RNAs are found in Bacillota and Gammaproteobacteria.

The xerDC RNA motif is a conserved RNA structure that was discovered by bioinformatics. xerDC motif RNAs are found in Clostridia.















