
Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dimensional structural configuration. In contrast to genetics, which refers to the study of individual genes and their roles in inheritance, genomics aims at the collective characterization and quantification of all of an organism's genes, their interrelations and influence on the organism. Genes may direct the production of proteins with the assistance of enzymes and messenger molecules. In turn, proteins make up body structures such as organs and tissues as well as control chemical reactions and carry signals between cells. Genomics also involves the sequencing and analysis of genomes through uses of high throughput DNA sequencing and bioinformatics to assemble and analyze the function and structure of entire genomes. Advances in genomics have triggered a revolution in discovery-based research and systems biology to facilitate understanding of even the most complex biological systems such as the brain.

A DNA sequencer is a scientific instrument used to automate the DNA sequencing process. Given a sample of DNA, a DNA sequencer is used to determine the order of the four bases: G (guanine), C (cytosine), A (adenine) and T (thymine). This is then reported as a text string, called a read. Some DNA sequencers can be also considered optical instruments as they analyze light signals originating from fluorochromes attached to nucleotides.

Molecular genetics is a sub-field of biology that addresses how differences in the structures or expression of DNA molecules manifests as variation among organisms. Molecular genetics often applies an "investigative approach" to determine the structure and/or function of genes in an organism's genome using genetic screens. The field of study is based on the merging of several sub-fields in biology: classical Mendelian inheritance, cellular biology, molecular biology, biochemistry, and biotechnology. Researchers search for mutations in a gene or induce mutations in a gene to link a gene sequence to a specific phenotype. Molecular genetics is a powerful methodology for linking mutations to genetic conditions that may aid the search for treatments/cures for various genetics diseases.

DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.

Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Frederick Sanger and colleagues in 1977, it became the most widely used sequencing method for approximately 40 years. It was first commercialized by Applied Biosystems in 1986. More recently, higher volume Sanger sequencing has been replaced by next generation sequencing methods, especially for large-scale, automated genome analyses. However, the Sanger method remains in wide use for smaller-scale projects and for validation of deep sequencing results. It still has the advantage over short-read sequencing technologies in that it can produce DNA sequence reads of > 500 nucleotides and maintains a very low error rate with accuracies around 99.99%. Sanger sequencing is still actively being used in efforts for public health initiatives such as sequencing the spike protein from SARS-CoV-2 as well as for the surveillance of norovirus outbreaks through the Center for Disease Control and Prevention's (CDC) CaliciNet surveillance network.

Jozef Stefaan "Jeff", Baron Schell was a Belgian molecular biologist.

2'-5'-oligoadenylate synthetase 1 is an enzyme that in humans is encoded by the OAS1 gene.

Homeobox D10, also known as HOXD10, is a protein which in humans is encoded by the HOXD10 gene.
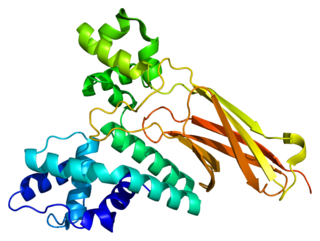
U5 small nuclear ribonucleoprotein 200 kDa helicase is an enzyme that in humans is encoded by the SNRNP200 gene.

Cleavage and polyadenylation specificity factor subunit 2 is a protein that in humans is encoded by the CPSF2 gene. This protein is a subunit of the cleavage and polyadenylation specificity factor (CPSF) complex which plays a key role in pre-mRNA 3' end processing and polyadenylation.The CPSF2 protein connects the two subunits of the complex, mCF and mPSF. Its structure contributes both to the stability of the subunits interaction and to the flexibility of the complex necessary for function. This protein has been identified as an essential subunit of the complex as certain mutations in the region inhibit CPSF complex formation.

Zinc finger protein 43 is a protein that in humans is encoded by the ZNF43 gene.

Poly(rC)-binding protein 3 is a protein that in humans is encoded by the PCBP3 gene.
RNA, U2 small nuclear, also known as RNU2, is a human gene.

snRNA-activating protein complex subunit 1 is a protein that in humans is encoded by the SNAPC1 gene.

Forkhead box protein D4 is a protein that in humans is encoded by the FOXD4 gene.

Calcyphosin is a protein that in humans is encoded by the CAPS gene.

GPI mannosyltransferase 3 is an enzyme that in humans is encoded by the PIGB gene.
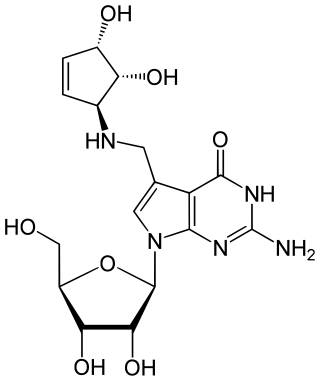
Queuosine is a modified nucleoside that is present in certain tRNAs in bacteria and eukaryotes. It contains the nucleobase queuine. Originally identified in E. coli, queuosine was found to occupy the first anticodon position of tRNAs for histidine, aspartic acid, asparagine and tyrosine. The first anticodon position pairs with the third "wobble" position in codons, and queuosine improves accuracy of translation compared to guanosine. Synthesis of queuosine begins with GTP. In bacteria, three structurally unrelated classes of riboswitch are known to regulate genes that are involved in the synthesis or transport of pre-queuosine1, a precursor to queuosine: PreQ1-I riboswitches, PreQ1-II riboswitches and PreQ1-III riboswitches.

Poly(A) polymerase gamma is an enzyme that in humans is encoded by the PAPOLG gene.

Riccardo Cortese was an Italian scientist, entrepreneur, and innovator in the field of gene expression, drug discovery and genetic vaccines. His work led to the development of novel therapeutic strategies for the prevention and cure of viral infections, including HIV, HCV, Ebola and RSV. He pioneered a novel platform technology based on simian adenoviral vectors for prophylactic and therapeutic vaccines, and authored more than 300 publications in peer reviewed journals in the field of gene expression, transcriptional control, molecular virology and immunology.














