Related Research Articles

The ykkC/yxkD leader is a conserved RNA structure found upstream of the ykkC and yxkD genes in Bacillus subtilis and related genes in other bacteria. The function of this family is unclear for many years although it has been suggested that it may function to switch on efflux pumps and detoxification systems in response to harmful environmental molecules. The Thermoanaerobacter tengcongensis sequence AE013027 overlaps with that of purine riboswitch suggesting that the two riboswitches may work in conjunction to regulate the upstream gene which codes for TTE0584 (Q8RC62), a member of the permease family.

6C RNA is a class of non-coding RNA present in actinomycetes. 6C RNA was originally discovered as a conserved RNA structure having two stem-loops each containing six or more cytosine (C) residues. Later work revealed that 6C RNAs in Streptomyces coelicolor and Streptomyces avermitilis have predicted rho-independent transcription terminators, and microarray and reverse-transcriptase PCR experiments indicate that the S. coelicolor version is transcribed as RNA. Transcription of the S. coelicolor RNA increases during sporulation, and three transcripts were detected that overlap the 6C motif, but have different apparent start and stop sites.
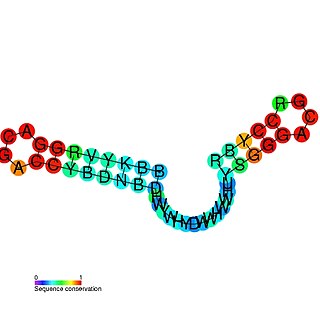
The mini-ykkC RNA motif was discovered as a putative RNA structure that is conserved in bacteria. The motif consists of two conserved stem-loops whose terminal loops contain the RNA sequence ACGR, where R represents either A or G. Mini-ykkC RNAs are widespread in Proteobacteria, but some are predicted in other phyla of bacteria. It was expected that the RNAs are cis-regulatory elements, because they are typically located upstream of protein-coding genes.

The ATPC RNA motif is a conserved RNA structure found in certain cyanobacteria. It is apparently ubiquitous in Prochlorococcus marinus, and is present in many species in the genus Synechococcus. The RNA is always found within an operon encoding subunits of ATP synthase, and it is always located downstream of the gene encoding the A subunit of ATP synthase, and upstream of the C subunit gene. This location is consistent with a cis-regulatory element, but also with a non-coding RNA that is transcribed with the ATP synthase genes.
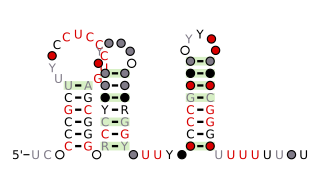
The anti-hemB RNA motif is a conserved RNA structure that was found in all known bacteria in the genus Burkholderia, and in a variety of other betaproteobacteria. The anti-hemB RNA motif consists primarily of two stem-loops, followed by a predicted rho-independent transcription termination stem-loop. As anti-hemB RNAs are generally not located in a 5' UTR, the RNAs are presumed to be non-coding RNAs. The terminator stem-loop implies that anti-hemB RNAs are transcribed as independent molecules.
The lacto-2 RNA motif is an RNA structure that is conserved amongst bacteria within the order Lactobacillales. The motif consists of a stem-loop whose stem is interrupted by many internal loops and bulges. Nucleotide identities in many places are conserved, and one internal loop in particular is highly conserved.
The MAEB RNA motif is a conserved stem-loop RNA structure present in many species in the genus Burkholderia. MAEB stem-loops typically occur in blocks of repeats, usually with 2–6 consecutive instances of MAEB stem-loops separated by a short and conserved linker sequence. As many as 12 consecutive MAEB stem-loops have been observed in a single block.

The Chlorobi-1 RNA motif is a conserved RNA secondary structure identified by bioinformatics. It is predicted to be used only by Chlorobi, a phylum of bacteria. The motif consists of two stem-loops that are followed by an apparent rho-independent transcription terminator. The motif is presumed to function as an independently transcribed non-coding RNA.

The Cyano-2 RNA motif is a conserved RNA structure identified by bioinformatics. Cyano-2 RNAs are found in Cyanobacterial species classified within the genus Synechococcus. Many terminal loops in the two conserved stem-loops contain the nucleotide sequence GCGA, and these sequences might in some cases form stable GNRA tetraloops. Since the two stem-loops are somewhat distant from one another it is possible that they represent two independent non-coding RNAs that are often or always co-transcribed. The region one thousand base pairs upstream of predicted Cyano-2 RNAs is usually devoid of annotated features such as RNA or protein-coding genes. This absence of annotated genes within one thousand base pairs is relatively unusual within bacteria.

The gabT RNA motif is the name of a conserved RNA structure identified by bioinformatics whose function is unknown. The gabT motif has been detected exclusively in bacteria within the genus Pseudomonas, and is found only upstream of gabT genes, and downstream to gabD genes.

The gyrA RNA motif is a conserved RNA structure identified by bioinformatics. The RNAs are present in multiple species of bacteria within the order Pseudomonadales. This order contains the genus Pseudomonas, which includes the opportunistic human pathogen Pseudomonas aeruginosa and Pseudomonas syringae, a plant pathogen.

The livK RNA motif describes a conserved RNA structured that was discovered using bioinformatics. The livK motif is detected only in the species Pseudomonas syringae. It is found in the potential 5' untranslated regions of livK genes and downstream livM and livH genes, as well as the 5' UTRs of amidase genes. The liv genes are predicted to be transporters of branched-chain amino acids, i.e., leucine, isoleucine or valine. The specific reaction catalyzed the amidase genes is not predicted.

The mraW RNA motif is a conserved, structured RNA found in certain bacteria. Specifically, it is predicted in many, though not all, species of actinobacteria, and especially within the genus Mycobacterium. Structurally, the motif consists of a hairpin with a highly conserved terminal loop sequence. mraW RNAs are consistently in the presumed 5' untranslated regions of mraW genes. These mraW genes likely form operons with immediately downstream ftsI genes, and multiple types of mur genes. These genes are associated with peptidoglycan synthesis, and it was hypothesized that the mraW RNA motif might regulate these genes.

The pan RNA motif defines a conserved RNA structure that was identified using bioinformatics. pan motif RNAs are present in three phyla: Chloroflexi, Firmicutes and Proteobacteria, although within the latter phylum they are only known in deltaproteobacteria. A pan RNA is present in the Firmicute Bacillus subtilis, which is one of the most extensively studied bacteria.

The psaA RNA motif describes a class of RNAs with a common secondary structure. psaA RNAs are exclusively found in locations that presumably correspond to the 5' untranslated regions of operons formed of psaA and psaB genes. For this reason, it was hypothesized that psaA RNAs function as cis-regulatory elements of these genes. The psaAB genes encode proteins that form subunits in the photosystem I structure used for photosynthesis. psaA RNAs have been detected only in cyanobacteria, which is consistent with their association with photosynthesis.
The Pseudomon-groES RNA motif is a conserved RNA structure identified in certain bacteria using bioinformatics. It is found in most species within the family Pseudomonadaceae, and is consistently located in the 5' untranslated regions of operons that contain groES genes. RNA transcripts of the groES genes in Pseudomonas aeruginosa where shown experimentally to be initiated at one of two start sites, from promoters called "P1" and "P2". The Pseudomon-groES RNA is in the 5' UTR of transcripts initiated from the P1 site, but is truncated in P2 transcripts. groES genes are involved in the cellular response to heat shock, but it is not thought that the Pseudomonas-groES RNA motif is involved in heat shock regulation. However, it is thought that the motif might regulate groES genes in response to other stimuli.
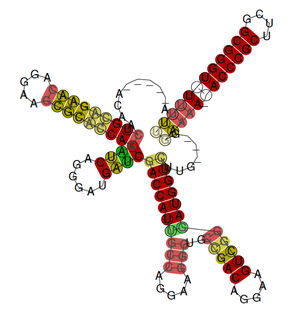
The rsmX gene is part of the Rsm/Csr family of non-coding RNAs (ncRNAs). Members of the Rsm/Csr family are present in a diverse range of bacteria, including Escherichia coli, Erwinia, Salmonella, Vibrio and Pseudomonas. These ncRNAs act by sequestering translational repressor proteins, called RsmA, activating expression of downstream genes that would normally be blocked by the repressors. Sequestering of target proteins is dependent upon exposed GGA motifs in the stem loops of the ncRNAs. Typically, the activated genes are involved in secondary metabolism, biofilm formation and motility.

The TwoAYGGAY RNA motif is a conserved RNA structure identified by bioinformatics. Its name refers to the conserved AYGGAY nucleotide sequence found in the motif's two terminal loops. The RNAs are found in sequences derived from DNA extracted from uncultivated bacteria present in the human gut, as well as some bacteria in the classes Clostridia and Gammaproteobacteria.
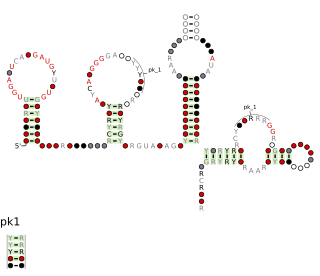
The Plasmid-Associated gamma-Proteobacteria Especially Vibrionales RNA motif is a conserved RNA structure that was discovered by bioinformatics. PAGEV motif RNAs are found in Gammaproteobacteria, especially within the order Vibrionales.

The Zeta-pan RNA motif is a conserved RNA structure that was discovered by bioinformatics. Zeta-pan motif RNAs are found in Zetaproteobacteria.
References
- ↑ Weinberg Z, Barrick JE, Yao Z, et al. (2007). "Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline". Nucleic Acids Res. 35 (14): 4809–4819. doi:10.1093/nar/gkm487. PMC 1950547 . PMID 17621584.
- ↑ Filiatrault MJ, Stodghill PV, Bronstein PA, et al. (May 2010). "Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity". J. Bacteriol. 192 (9): 2359–2372. doi:10.1128/JB.01445-09. PMC 2863471 . PMID 20190049.
| This molecular or cell biology article is a stub. You can help Wikipedia by expanding it. |