Related Research Articles

An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.
Gene knockouts are a widely used genetic engineering technique that involves the targeted removal or inactivation of a specific gene within an organism's genome. This can be done through a variety of methods, including homologous recombination, CRISPR-Cas9, and TALENs.

The Philadelphia chromosome or Philadelphia translocation (Ph) is a specific genetic abnormality in chromosome 22 of leukemia cancer cells. This chromosome is defective and unusually short because of reciprocal translocation, t(9;22)(q34;q11), of genetic material between chromosome 9 and chromosome 22, and contains a fusion gene called BCR-ABL1. This gene is the ABL1 gene of chromosome 9 juxtaposed onto the breakpoint cluster region BCR gene of chromosome 22, coding for a hybrid protein: a tyrosine kinase signaling protein that is "always on", causing the cell to divide uncontrollably by interrupting the stability of the genome and impairing various signaling pathways governing the cell cycle.

Chronic myelogenous leukemia (CML), also known as chronic myeloid leukemia, is a cancer of the white blood cells. It is a form of leukemia characterized by the increased and unregulated growth of myeloid cells in the bone marrow and the accumulation of these cells in the blood. CML is a clonal bone marrow stem cell disorder in which a proliferation of mature granulocytes and their precursors is found; characteristic increase in basophils is clinically relevant. It is a type of myeloproliferative neoplasm associated with a characteristic chromosomal translocation called the Philadelphia chromosome.
The Cancer Genome Project is part of the cancer, aging, and somatic mutation research based at the Wellcome Trust Sanger Institute in the United Kingdom. It aims to identify sequence variants/mutations critical in the development of human cancers. Like The Cancer Genome Atlas project within the United States, the Cancer Genome Project represents an effort in the War on Cancer to improve cancer diagnosis, treatment, and prevention through a better understanding of the molecular basis of the disease. The Cancer Genome Project was launched by Michael Stratton in 2000, and Peter Campbell is now the group leader of the project. The project works to combine knowledge of the human genome sequence with high throughput mutation detection techniques.

Chemokine ligand 9 (CCL9) is a small cytokine belonging to the CC chemokine family. It is also called macrophage inflammatory protein-1 gamma (MIP-1γ), macrophage inflammatory protein-related protein-2 (MRP-2) and CCF18, that has been described in rodents. CCL9 has also been previously designated CCL10, although this name is no longer in use. It is secreted by follicle-associated epithelium (FAE) such as that found around Peyer's patches, and attracts dendritic cells that possess the cell surface molecule CD11b and the chemokine receptor CCR1. CCL9 can activate osteoclasts through its receptor CCR1 suggesting an important role for CCL9 in bone resorption. CCL9 is constitutively expressed in macrophages and myeloid cells. The gene for CCL9 is located on chromosome 11 in mice.

Acute myeloblastic leukemia with maturation (M2) is a subtype of acute myeloid leukemia (AML).

Wilms tumor protein (WT33) is a protein that in humans is encoded by the WT1 gene on chromosome 11p.

CCAAT/enhancer-binding protein alpha is a protein encoded by the CEBPA gene in humans. CCAAT/enhancer-binding protein alpha is a transcription factor involved in the differentiation of certain blood cells. For details on the CCAAT structural motif in gene enhancers and on CCAAT/Enhancer Binding Proteins see the specific page.

Factor interacting with PAPOLA and CPSF1 is a protein that in humans is encoded by the FIP1L1 gene. A medically important aspect of the FIP1L1 gene is its fusion with other genes to form fusion genes which cause clonal hypereosinophilia and leukemic diseases in humans.

Acute megakaryoblastic leukemia (AMKL) is life-threatening leukemia in which malignant megakaryoblasts proliferate abnormally and injure various tissues. Megakaryoblasts are the most immature precursor cells in a platelet-forming lineage; they mature to promegakaryocytes and, ultimately, megakaryocytes which cells shed membrane-enclosed particles, i.e. platelets, into the circulation. Platelets are critical for the normal clotting of blood. While malignant megakaryoblasts usually are the predominant proliferating and tissue-damaging cells, their similarly malignant descendants, promegakaryocytes and megakaryocytes, are variable contributors to the malignancy.

In molecular biology, mutagenesis is an important laboratory technique whereby DNA mutations are deliberately engineered to produce libraries of mutant genes, proteins, strains of bacteria, or other genetically modified organisms. The various constituents of a gene, as well as its regulatory elements and its gene products, may be mutated so that the functioning of a genetic locus, process, or product can be examined in detail. The mutation may produce mutant proteins with interesting properties or enhanced or novel functions that may be of commercial use. Mutant strains may also be produced that have practical application or allow the molecular basis of a particular cell function to be investigated.
KBM-7 cells are a chronic myelogenous leukemia (CML) cell line used for biomedical research. Like all cancer cell lines, it is immortal and can divide indefinitely. A unique aspect of the KBM-7 cell line is that it is near-haploid, meaning it contains only one copy for most of its chromosomes. Human chromosomes are typically diploid, meaning that there are two copies of each chromosome.
AI-10-49 is a small molecule inhibitor of leukemic oncoprotein CBFβ-SMHHC developed by the laboratory of John Bushweller with efficacy demonstrated by the laboratories of Lucio H. Castilla and Monica Guzman. AI-10-49 allosterically binds to CBFβ-SMMHC and disrupts protein-protein interaction between CBFβ-SMMHC and tumor suppressor RUNX1. This inhibitor is under development as an anti-leukemic drug.

Musashi-2, also known as Musashi RNA binding protein 2, is a protein that in humans is encoded by the MSI2 gene. Like its homologue musashi-1 (MSI1), it is an RNA-binding protein involved in stemness.
Human germline engineering is the process by which the genome of an individual is edited in such a way that the change is heritable. This is achieved by altering the genes of the germ cells, which then mature into genetically modified eggs and sperm. For safety, ethical, and social reasons, there is broad agreement among the scientific community and the public that germline editing for reproduction is a red line that should not be crossed at this point in time. There are differing public sentiments, however, on whether it may be performed in the future depending on whether the intent would be therapeutic or non-therapeutic.
Off-target genome editing refers to nonspecific and unintended genetic modifications that can arise through the use of engineered nuclease technologies such as: clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9, transcription activator-like effector nucleases (TALEN), meganucleases, and zinc finger nucleases (ZFN). These tools use different mechanisms to bind a predetermined sequence of DNA (“target”), which they cleave, creating a double-stranded chromosomal break (DSB) that summons the cell's DNA repair mechanisms and leads to site-specific modifications. If these complexes do not bind at the target, often a result of homologous sequences and/or mismatch tolerance, they will cleave off-target DSB and cause non-specific genetic modifications. Specifically, off-target effects consist of unintended point mutations, deletions, insertions inversions, and translocations.

CRISPR gene editing is a genetic engineering technique in molecular biology by which the genomes of living organisms may be modified. It is based on a simplified version of the bacterial CRISPR-Cas9 antiviral defense system. By delivering the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) into a cell, the cell's genome can be cut at a desired location, allowing existing genes to be removed and/or new ones added in vivo.
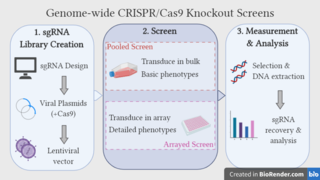
Genome-wide CRISPR-Cas9 knockout screens aim to elucidate the relationship between genotype and phenotype by ablating gene expression on a genome-wide scale and studying the resulting phenotypic alterations. The approach utilises the CRISPR-Cas9 gene editing system, coupled with libraries of single guide RNAs (sgRNAs), which are designed to target every gene in the genome. Over recent years, the genome-wide CRISPR screen has emerged as a powerful tool for performing large-scale loss-of-function screens, with low noise, high knockout efficiency and minimal off-target effects.
Nissim Benvenisty is Professor of Genetics, the Herbert Cohn Chair in Cancer Research and the Director of “The Azrieli Center for Stem Cells and Genetic Research” at the Alexander Silberman Institute of Life Sciences, Hebrew University.
References
- ↑ "HAP1 Cells". www.horizondiscovery.com. Retrieved 2016-04-14.
- ↑ Carette, Jan E.; Guimaraes, Carla P.; Varadarajan, Malini; Park, Annie S.; Wuethrich, Irene; Godarova, Alzbeta; Kotecki, Maciej; Cochran, Brent H.; Spooner, Eric (2009-11-27). "Haploid Genetic Screens in Human Cells Identify Host Factors Used by Pathogens". Science. 326 (5957): 1231–1235. Bibcode:2009Sci...326.1231C. doi:10.1126/science.1178955. ISSN 0036-8075. PMID 19965467. S2CID 5287138.
- ↑ Kotecki, Maciej; Reddy, P. Sanjeeva; Cochran, Brent H. (1999-11-01). "Isolation and Characterization of a Near-Haploid Human Cell Line". Experimental Cell Research. 252 (2): 273–280. CiteSeerX 10.1.1.24.783 . doi:10.1006/excr.1999.4656. PMID 10527618.
- 1 2 3 Essletzbichler, Patrick; Konopka, Tomasz; Santoro, Federica; Chen, Doris; Gapp, Bianca V.; Kralovics, Robert; Brummelkamp, Thijn R.; Nijman, Sebastian M. B.; Bürckstümmer, Tilmann (2014-12-01). "Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line". Genome Research. 24 (12): 2059–2065. doi:10.1101/gr.177220.114. ISSN 1088-9051. PMC 4248322 . PMID 25373145.
- 1 2 3 4 Wutz, Anton (2014-04-01). "Haploid animal cells" (PDF). Development. 141 (7): 1423–1426. doi: 10.1242/dev.102202 . ISSN 0950-1991. PMID 24644259.
- 1 2 Oshimura, Mitsuo; Freeman, Arnold I.; Sandberg, Avery A. (1977-09-01). "Chromosomes and causation of human cancer and leukemia. XXIII. Near-haploidy in acute leukemia". Cancer. 40 (3): 1143–1148. doi:10.1002/1097-0142(197709)40:3<1143::aid-cncr2820400325>3.0.co;2-4. ISSN 1097-0142. PMID 268995.
- 1 2 Baba, Alecsandru Ioan; Câtoi, Cornel (2007-01-01). "TUMOR CELL MORPHOLOGY". The Publishing House of the Romanian Academy.
{{cite journal}}: Cite journal requires|journal=(help) - ↑ "What is chronic myeloid leukemia?". www.cancer.org. Retrieved 2016-04-14.