Related Research Articles
Histocompatibility, or tissue compatibility, is the property of having the same, or sufficiently similar, alleles of a set of genes called human leukocyte antigens (HLA), or major histocompatibility complex (MHC). Each individual expresses many unique HLA proteins on the surface of their cells, which signal to the immune system whether a cell is part of the self or an invading organism. T cells recognize foreign HLA molecules and trigger an immune response to destroy the foreign cells. Histocompatibility testing is most relevant for topics related to whole organ, tissue, or stem cell transplants, where the similarity or difference between the donor's HLA alleles and the recipient's triggers the immune system to reject the transplant. The wide variety of potential HLA alleles lead to unique combinations in individuals and make matching difficult.

The major histocompatibility complex (MHC) is a large locus on vertebrate DNA containing a set of closely linked polymorphic genes that code for cell surface proteins essential for the adaptive immune system. These cell surface proteins are called MHC molecules.

The human leukocyte antigen (HLA) system is a complex of genes on chromosome 6 in humans that encode cell-surface proteins responsible for regulation of the immune system. The HLA system is also known as the human version of the major histocompatibility complex (MHC) found in many animals.
In academia, computational immunology is a field of science that encompasses high-throughput genomic and bioinformatics approaches to immunology. The field's main aim is to convert immunological data into computational problems, solve these problems using mathematical and computational approaches and then convert these results into immunologically meaningful interpretations.
HLA-DR is an MHC class II cell surface receptor encoded by the human leukocyte antigen complex on chromosome 6 region 6p21.31. The complex of HLA-DR and peptide, generally between 9 and 30 amino acids in length, constitutes a ligand for the T-cell receptor (TCR). HLA were originally defined as cell surface antigens that mediate graft-versus-host disease. Identification of these antigens has led to greater success and longevity in organ transplant.

HLA class II histocompatibility antigen, DRB1 beta chain is a protein that in humans is encoded by the HLA-DRB1 gene. DRB1 encodes the most prevalent beta subunit of HLA-DR. DRB1 alleles, especially those encoding amino acid sequence changes at positions 11 and 13, are associated risk of rheumatoid arthritis.
HLA-DQ (DQ) is a cell surface receptor protein found on antigen-presenting cells. It is an αβ heterodimer of type MHC class II. The α and β chains are encoded by two loci, HLA-DQA1 and HLA-DQB1, that are adjacent to each other on chromosome band 6p21.3. Both α-chain and β-chain vary greatly. A person often produces two α-chain and two β-chain variants and thus 4 isoforms of DQ. The DQ loci are in close genetic linkage to HLA-DR, and less closely linked to HLA-DP, HLA-A, HLA-B and HLA-C.
HLA-DP is a protein/peptide-antigen receptor and graft-versus-host disease antigen that is composed of 2 subunits, DPα and DPβ. DPα and DPβ are encoded by two loci, HLA-DPA1 and HLA-DPB1, that are found in the MHC Class II region in the Human Leukocyte Antigen complex on human chromosome 6 . Less is known about HLA-DP relative to HLA-DQ and HLA-DR but the sequencing of DP types and determination of more frequent haplotypes has progressed greatly within the last few years.
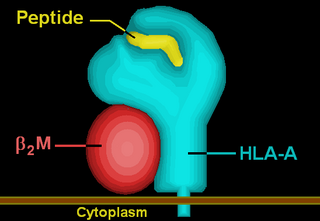
HLA-A is a group of human leukocyte antigens (HLA) that are encoded by the HLA-A locus, which is located at human chromosome 6p21.3. HLA is a major histocompatibility complex (MHC) antigen specific to humans. HLA-A is one of three major types of human MHC class I transmembrane proteins. The others are HLA-B and HLA-C. The protein is a heterodimer, and is composed of a heavy α chain and smaller β chain. The α chain is encoded by a variant HLA-A gene, and the β chain (β2-microglobulin) is an invariant β2 microglobulin molecule. The β2 microglobulin protein is encoded by the B2M gene, which is located at chromosome 15q21.1 in humans.
Tissue typing is a procedure in which the tissues of a prospective donor and recipient are tested for compatibility prior to transplantation. Mismatched donor and recipient tissues can lead to rejection of the tissues. There are multiple methods of tissue typing.
Immunogenetics or immungenetics is the branch of Medical Immunology and Medical Genetics that explores the relationship between the immune system and genetics.

HLA class II histocompatibility antigen, DRB5 beta chain is a protein that in humans is encoded by the HLA-DRB5 gene.
HLA-DR5 (DR5) is a broad-antigen serotype that is further split into HLA-DR11 and HLA-DR12 antigen serotypes.

HLA-A*02 (A*02) is a human leukocyte antigen serotype within the HLA-A serotype group. The serotype is determined by the antibody recognition of the α2 domain of the HLA-A α-chain. For A*02, the α chain is encoded by the HLA-A*02 gene and the β chain is encoded by the B2M locus. In 2010 the World Health Organization Naming Committee for Factors of the HLA System revised the nomenclature for HLAs. Before this revision, HLA-A*02 was also referred to as HLA-A2, HLA-A02, and HLA-A*2.
Major histocompatibility complex, class II, DQ alpha 1, also known as HLA-DQA1, is a human gene present on short arm of chromosome 6 (6p21.3) and also denotes the genetic locus which contains this gene. The protein encoded by this gene is one of two proteins that are required to form the DQ heterodimer, a cell surface receptor essential to the function of the immune system.
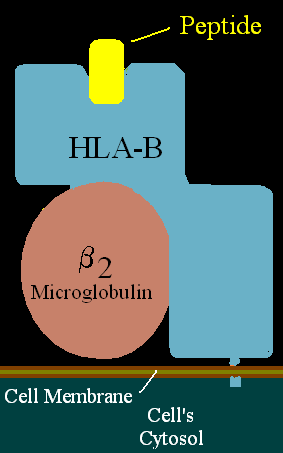
HLA-B7 (B7) is an HLA-B serotype. The serotype identifies the more common HLA-B*07 gene products. B7, previously HL-A7, was one of the first 'HL-A' antigens recognized, largely because of the frequency of B*0702 in Northern and Western Europe and the United States. B7 is found in two major haplotypes in Europe, where it reaches peak frequency in Ireland. One haplotype A3-B7-DR15-DQ1 can be found over a vast region and is in apparent selective disequilibrium. B7 is a risk factor for cervical cancer, sarcoidosis, and early-onset spondylarthropathies.

Enteropathy-associated T-cell lymphoma (EATL), previously termed enteropathy-associated T-cell lymphoma, type I and at one time termed enteropathy-type T-cell lymphoma (ETTL), is a complication of coeliac disease in which a malignant T-cell lymphoma develops in areas of the small intestine affected by the disease's intense inflammation. While a relatively rare disease, it is the most common type of primary gastrointestinal T-cell lymphoma.
Human leukocyte antigens (HLA) began as a list of antigens identified as a result of transplant rejection. The antigens were initially identified by categorizing and performing massive statistical analyses on interactions between blood types. This process is based upon the principle of serotypes. HLA are not typical antigens, like those found on surface of infectious agents. HLAs are alloantigens, they vary from individual to individual as a result of genetic differences. An organ called the thymus is responsible for ensuring that any T-cells that attack self proteins are not allowed to live. In essence, every individual's immune system is tuned to the specific set of HLA and self proteins produced by that individual; where this goes awry is when tissues are transferred to another person. Since individuals almost always have different "banks" of HLAs, the immune system of the recipient recognizes the transplanted tissue as non-self and destroys the foreign tissue, leading to transplant rejection. It was through the realization of this that HLAs were discovered.
Steven GE Marsh is a British Immunogeneticist and leader in the field of histocompatibility and immunogenetics having published more than 400 scientific papers on the subject. Marsh is Professor of Immunogenetics within University College London and the Chief Bioinformatics and Immunogenetics Officer at the charity Anthony Nolan where his heads the HLA Informatics Group.
IMGT or the international ImMunoGeneTics information system is a collection of databases and resources for immunoinformatics, particularly the V, D, J, and C gene sequences, as well as a providing other tools and data related to the adaptive immune system. IMGT/LIGM-DB, the first and still largest database hosted as part of IMGT contains reference nucleotide sequences for 360 species' T-cell receptor and immunoglobulin molecules, as of 2023. These genes encode the proteins which are the foundation of adaptive immunity, which allows highly specific recognition and memory of pathogens.
References
- ↑ Robinson, James; Barker, Dominic J; Georgiou, Xenia; Cooper, Michael A; Flicek, Paul; Marsh, Steven G E (2020). "IPD-IMGT/HLA Database". Nucleic Acids Research. 48 (D1): D948–D955. doi:10.1093/nar/gkz950. PMC 7145640 . PMID 31667505.
- ↑ About the Databases
- ↑ "HLA Informatics Group". www.anthonynolan.org. Retrieved 2024-11-17.