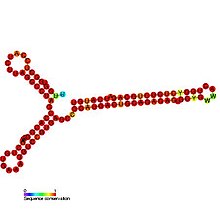
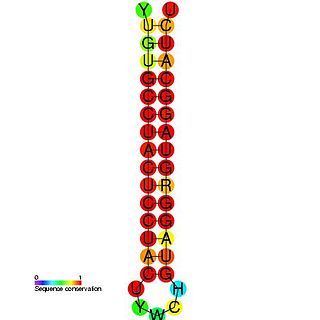
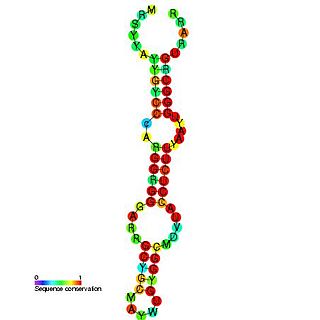
Hepatitis D is a disease caused by the hepatitis D virus (HDV), a small spherical enveloped virusoid. This is one of five known hepatitis viruses: A, B, C, D, and E. HDV is considered to be a subviral satellite because it can propagate only in the presence of the hepatitis B virus (HBV). Transmission of HDV can occur either via simultaneous infection with HBV (coinfection) or superimposed on chronic hepatitis B or hepatitis B carrier state (superinfection).

Hepadnaviridae is a family of viruses. Humans, apes, and birds serve as natural hosts. There are currently seven species in this family, divided among 2 genera. Its best-known member is the hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas, and cirrhosis.

Hepatitis C virus (HCV), a member of the Hepacivirus C species, is a small, enveloped, positive-sense single-stranded RNA virus of the family Flaviviridae. The hepatitis C virus is the cause of hepatitis C and some cancers such as liver cancer and lymphomas in humans.

Rubella virus (RuV) is the pathogenic agent of the disease rubella, and is the cause of congenital rubella syndrome when infection occurs during the first weeks of pregnancy.

The Coronavirus packaging signal is a conserved cis-regulatory element found in Coronavirus which has an important role in regulating the packaging of the viral genome into the capsid.

The coronavirus SL-III cis-acting replication element (CRE) is an RNA element that regulates defective interfering (DI) RNA replication.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.

The hepatitis E virus cis-reactive element is a RNA element that is thought to be essential for "some step in gene expression". The mutation of this element resulted in hepatitis E strains which were unable to infect rhesus macaques.

Retroviral Psi packaging element is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.

The Human Parechovirus 1 cis regulatory element is an RNA element which is located in the 5'-terminal 112 nucleotides of the genome of human parechovirus 1 (HPeV1). The element consists of two stem-loop structures together with a pseudoknot. Disruption of any of these elements impairs both viral replication and growth.
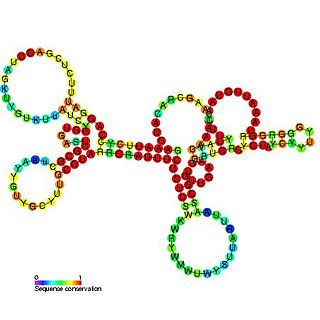
Tombusvirus 5' UTR is an important cis-regulatory region of the Tombus virus genome.
The TCV hairpin 5 (H5) is an RNA element found in the turnip crinkle virus. This RNA element is composed of a stem-loop that contains a large symmetrical internal loop (LSL). H5 can repress minus-strand synthesis when the 3' side of the LSL pairs with the 4 bases at the 3'-terminus of the RNA(GCCC-OH).
NSP1, the product of rotavirus gene 5, is a nonstructural RNA-binding protein that contains a cysteine-rich region and is a component of early replication intermediates. RNA-folding predictions suggest that this region of the NSP1 mRNA can interact with itself, producing a stem-loop structure similar to that found near the 5'-terminus of the NSP1 mRNA.
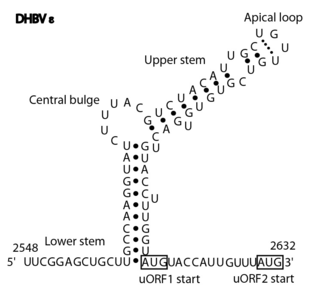
The Avian HBV RNA encapsidation signal epsilon is an RNA structure that is shown to facilitate encapsidation of the pregenomic RNA required for replication. There are two main classes of encapsidation signals in avian HBV viruses - Duck hepatitis B virus (DHBV) and Heron hepatitis B virus (HHBV) like. DHBV is used as a model to understand human HBV. Although studies have shown that the HHBV epsilon has less pairing in the upper stem than DHBV, this pairing is not absolutely required for DHBV infection in ducks.

Hepatitis B virus, abbreviated HBV, is a partially double-stranded DNA virus, a species of the genus Orthohepadnavirus and a member of the Hepadnaviridae family of viruses. This virus causes the disease hepatitis B.
In molecular biology, the Hepatitis A virus cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Hepatitis A virus (HAV). It is larger than the CREs found in related Picornavirus species, but is thought to be functionally similar. It is thought to be involved in uridylylation of VPg.
In molecular biology, the Avian encephalitis virus cis-acting replication element (CRE) is an s an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Avian encephalitis virus (AEV). It is structurally similar to the Hepatitis A virus cis-acting replication element.