Related Research Articles

A bacteriophage, also known informally as a phage, is a virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.
Bacterial conjugation is the transfer of genetic material between bacterial cells by direct cell-to-cell contact or by a bridge-like connection between two cells. This takes place through a pilus.

An endosymbiont or endobiont is any organism that lives within the body or cells of another organism most often, though not always, in a mutualistic relationship. (The term endosymbiosis is from the Greek: ἔνδον endon "within", σύν syn "together" and βίωσις biosis "living".) Examples are nitrogen-fixing bacteria, which live in the root nodules of legumes; single-cell algae inside reef-building corals, and bacterial endosymbionts that provide essential nutrients to about 10–15% of insects.

In the fields of molecular biology and genetics, a genome is the genetic material of an organism. It consists of DNA. The genome includes both the genes and the noncoding DNA, as well as mitochondrial DNA and chloroplast DNA. The study of the genome is called genomics.

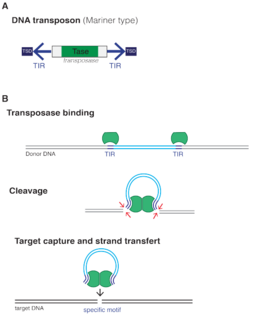
A transposable element is a DNA sequence that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Transposition often results in duplication of the same genetic material. Barbara McClintock's discovery of them earned her a Nobel Prize in 1983.

Escherichia coli, also known as E. coli, is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms (endotherms). Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. The harmless strains are part of the normal microbiota of the gut, and can benefit their hosts by producing vitamin K2, (which helps blood to clot) and preventing colonisation of the intestine with pathogenic bacteria, having a symbiotic relationship. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for 3 days, but its numbers decline slowly afterwards.

Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between unicellular and/or multicellular organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the evolution of many organisms.

The last universal common ancestor or last universal cellular ancestor (LUCA), also called the last universal ancestor (LUA), or concestor, is the most recent population of organisms from which all organisms now living on Earth have a common descent, the most recent common ancestor of all current life on Earth. LUCA is not thought to be the first life on Earth, but rather the only organism of its time to still have living descendants.

A tumour inducing (Ti) plasmid is a plasmid found in pathogenic species of Agrobacterium, including A. tumefaciens, A. rhizogenes, A. rubi and A. vitis.

In microbiology, genetics, cell biology, and molecular biology, competence is the ability of a cell to alter its genetics by taking up extracellular ("naked") DNA from its environment in the process called transformation. Competence may be differentiated between natural competence, a genetically specified ability of bacteria which is thought to occur under natural conditions as well as in the laboratory, and induced or artificial competence, which arises when cells in laboratory cultures are treated to make them transiently permeable to DNA. Competence allows for rapid adaptation and DNA repair of the cell. This article primarily deals with natural competence in bacteria, although information about artificial competence is also provided.

Mobile genetic elements (MGEs) are a type of genetic material that can move around within a genome, or that can be transferred from one species or replicon to another. MGEs are found in all organisms. In humans, approximately 50% of the genome is thought to be MGEs. MGEs play a distinct role in evolution. Gene duplication events can also happen through the mechanism of MGEs. MGEs can also cause mutations in protein coding regions, which alters the protein functions. They can also rearrange genes in the host genome. One of the examples of MGEs in evolutionary context is that virulence factors and antibiotic resistance genes of MGEs can be transported to share them with neighboring bacteria. Newly acquired genes through this mechanism can increase fitness by gaining new or additional functions. On the other hand, MGEs can also decrease fitness by introducing disease-causing alleles or mutations.
The mobilome is the entire set of mobile genetic elements in a genome. Mobilomes are found in eukaryotes, prokaryotes, and viruses. The compositions of mobilomes differ among lineages of life, with transposable elements being the major mobile elements in eukaryotes, and plasmids and prophages being the major types in prokaryotes. Virophages contribute to the viral mobilome.

16S ribosomal RNA is the component of the 30S small subunit of a prokaryotic ribosome that binds to the Shine-Dalgarno sequence. The genes coding for it are referred to as 16S rRNA gene and are used in reconstructing phylogenies, due to the slow rates of evolution of this region of the gene. Carl Woese and George E. Fox were two of the people who pioneered the use of 16S rRNA in phylogenetics in 1977.

A prokaryote is a unicellular organism that lacks a membrane-bound nucleus, mitochondria, or any other membrane-bound organelle. The word prokaryote comes from the Greek πρό and κάρυον. Prokaryotes are divided into two domains, Archaea and Bacteria. Organisms with nuclei and other organelles are placed in a third domain, Eukaryota. Prokaryotes are asexual, reproducing without fusion of gametes. The first organisms are thought to have been prokaryotes.

Archaea constitute a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria, but this classification is obsolete.

Scientists trying to reconstruct evolutionary history have been challenged by the fact that genes can sometimes transfer between distant branches on the tree of life. This movement of genes can occur through horizontal gene transfer (HGT), scrambling the information on which biologists relied to reconstruct the phylogeny of organisms. Conversely, HGT can also help scientists to reconstruct and date the tree of life. Indeed, a gene transfer can be used as a phylogenetic marker, or as the proof of contemporaneity of the donor and recipient organisms, and as a trace of extinct biodiversity.
Bacterial genomes are generally smaller and less variant in size among species when compared with genomes of eukaryotes. Bacterial genomes can range in size anywhere from about 130 kbp to over 14 Mbp. A study that included, but was not limited to, 478 bacterial genomes, concluded that as genome size increases, the number of genes increases at a disproportionately slower rate in eukaryotes than in non-eukaryotes. Thus, the proportion of non-coding DNA goes up with genome size more quickly in non-bacteria than in bacteria. This is consistent with the fact that most eukaryotic nuclear DNA is non-gene coding, while the majority of prokaryotic, viral, and organellar genes are coding. Right now, we have genome sequences from 50 different bacterial phyla and 11 different archaeal phyla. Second-generation sequencing has yielded many draft genomes ; third-generation sequencing might eventually yield a complete genome in a few hours. The genome sequences reveal much diversity in bacteria. Analysis of over 2000 Escherichia coli genomes reveals an E. coli core genome of about 3100 gene families and a total of about 89,000 different gene families. Genome sequences show that parasitic bacteria have 500–1200 genes, free-living bacteria have 1500–7500 genes, and archaea have 1500–2700 genes. A striking discovery by Cole et al. described massive amounts of gene decay when comparing Leprosy bacillus to ancestral bacteria. Studies have since shown that several bacteria have smaller genome sizes than their ancestors did. Over the years, researchers have proposed several theories to explain the general trend of bacterial genome decay and the relatively small size of bacterial genomes. Compelling evidence indicates that the apparent degradation of bacterial genomes is owed to a deletional bias.
The Eocyte hypothesis is a biological classification that indicates eukaryotes emerged within the prokaryotic Crenarchaeota, a phylum within the archaea. This hypothesis was originally proposed by James A. Lake and colleagues in 1984 based on the discovery that the shapes of ribosomes in the Crenarchaeota and eukaryotes are more similar to each other than to either bacteria or the second major kingdom of archaea, the Euryarchaeota.
Microbial phylogenetics is the study of the manner in which various groups of microorganisms are genetically related. This helps to trace their evolution. To study these relationships biologists rely on comparative genomics, as physiology and comparative anatomy are not possible methods.
Horizontal or lateral gene transfer is the transmission of portions of genomic DNA between organisms through a process decoupled from vertical inheritance. In the presence of HGT events, different fragments of the genome are the result of different evolutionary histories. This can therefore complicate the investigations of evolutionary relatedness of lineages and species. Also, as HGT can bring into genomes radically different genotypes from distant lineages, or even new genes bearing new functions, it is a major source of phenotypic innovation and a mechanism of niche adaptation. For example, of particular relevance to human health is the lateral transfer of antibiotic resistance and pathogenicity determinants, leading to the emergence of pathogenic lineages.
References
- ↑ Acuña, Ricardo; Padilla, Beatriz E.; Flórez-Ramos, Claudia P.; Rubio, José D.; Herrera, Juan C.; Benavides, Pablo; Lee, Sang-Jik; Yeats, Trevor H.; Egan, Ashley N.; Doyle, Jeffrey J.; Rose, Jocelyn K. C. (13 March 2012). "Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee". PNAS . 109 (11): 4197–4202. doi:10.1073/pnas.1121190109. PMC 3306691 . PMID 22371593.
- ↑ Phillips, Melissa Lee (27 February 2012). "Bacterial gene helps coffee beetle get its fix". Nature . doi:10.1038/nature.2012.10116.
| This gene article is a stub. You can help Wikipedia by expanding it. |