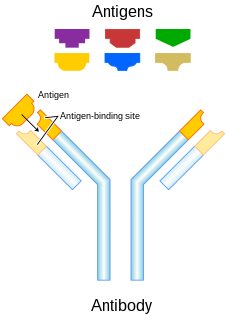
In immunology, antigens (Ag) are structures specifically bound by antibodies (Ab) or a cell surface version of Ab ~ B cell antigen receptor (BCR). The terms antigen originally described a structural molecule that binds specifically to an antibody only in the form of native antigen. It was expanded later to refer to any molecule or a linear molecular fragment after processing the native antigen that can be recognized by T-cell receptor (TCR). BCR and TCR are both highly variable antigen receptors diversified by somatic V(D)J recombination. Both T cells and B cells are cellular components of adaptive immunity. The Ag abbreviation stands for an antibody generator.

An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein produced mainly by plasma cells that is used by the immune system to neutralize pathogens such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen, via the fragment antigen-binding (Fab) variable region. Each tip of the "Y" of an antibody contains a paratope that is specific for one particular epitope on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can tag a microbe or an infected cell for attack by other parts of the immune system, or can neutralize its target directly. Depending on the antigen, the binding may impede the biological process causing the disease or may activate macrophages to destroy the foreign substance. The ability of an antibody to communicate with the other components of the immune system is mediated via its Fc region, which contains a conserved glycosylation site involved in these interactions. The production of antibodies is the main function of the humoral immune system.

Proteomics is the large-scale study of proteins. Proteins are vital parts of living organisms, with many functions. The term proteomics was coined in 1997, in analogy to genomics, the study of the genome. The word proteome is a portmanteau of protein and genome, and was coined by Marc Wilkins in 1994 while he was a Ph.D. student at Macquarie University. Macquarie University also founded the first dedicated proteomics laboratory in 1995.
Antigenic drift is a mechanism for variation in viruses that involves the accumulation of mutations within the genes that code for antibody-binding sites. This results in a new strain of virus particles which cannot be inhibited as effectively by the antibodies that were originally targeted against previous strains, making it easier for the virus to spread throughout a partially immune population. Antigenic drift occurs in both influenza A and influenza B viruses.
Virus-like particles resemble viruses, but are non-infectious because they contain no viral genetic material. The expression of viral structural proteins, such as envelope or capsid proteins, can result in the self-assembly of virus like particles (VLPs). VLPs derived from the Hepatitis B virus and composed of the small HBV derived surface antigen (HBsAg) were described in 1968 from patient sera. VLPs have been produced from components of a wide variety of virus families including Parvoviridae, Retroviridae, Flaviviridae and bacteriophages. VLPs can be produced in multiple cell culture systems including bacteria, mammalian cell lines, insect cell lines, yeast and plant cells.
Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds to that particular protein. This process can be used to isolate and concentrate a particular protein from a sample containing many thousands of different proteins. Immunoprecipitation requires that the antibody be coupled to a solid substrate at some point in the procedure.

Epitope mapping is the process of experimentally identifying the binding site, or "epitope", of an antibody on its target antigen. Identification and characterization of antibody binding sites aid in the discovery and development of new therapeutics, vaccines, and diagnostics. Epitope characterization can also help elucidate the mechanism of binding for an antibody and can strength intellectual property (patent) protection. Experimental epitope mapping data can be incorporated into robust algorithms to facilitate in silico prediction of B-cell epitopes based on sequence and/or structural data.

In mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy absorbing matrix to create ions from large molecules with minimal fragmentation. It has been applied to the analysis of biomolecules and large organic molecules, which tend to be fragile and fragment when ionized by more conventional ionization methods. It is similar in character to electrospray ionization (ESI) in that both techniques are relatively soft ways of obtaining ions of large molecules in the gas phase, though MALDI typically produces far fewer multi-charged ions.

A single-domain antibody (sdAb) is an antibody fragment consisting of a single monomeric variable antibody domain. Like a whole antibody, it is able to bind selectively to a specific antigen. With a molecular weight of only 12–15 kDa, single-domain antibodies are much smaller than common antibodies which are composed of two heavy protein chains and two light chains, and even smaller than Fab fragments and single-chain variable fragments.
Cross-reactivity, in a general sense, is the reactivity of an observed agent which initiates reactions outside the main reaction expected.
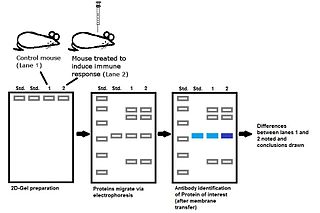
Immunoproteomics is the study of large sets of proteins (proteomics) involved in the immune response.
In biochemistry, avidity refers to the accumulated strength of multiple affinities of individual non-covalent binding interactions, such as between a protein receptor and its ligand, and is commonly referred to as functional affinity. As such, avidity is distinct from affinity, which describes the strength of a single interaction. However, because individual binding events increase the likelihood of other interactions to occur, avidity should not be thought of as the mere sum of its constituent affinities but as the combined effect of all affinities participating in the biomolecular interaction. A particular important aspect relates to the phenomenon of 'avidity entropy'. Biomolecules often form heterogenous complexes or homogeneous oligomers and multimers or polymers. If clustered proteins form an organized matrix, such as the clathrin-coat, the interaction is described as a matricity.
Surface-enhanced laser desorption/ionization (SELDI) is a soft ionization method in mass spectrometry (MS) used for the analysis of protein mixtures. It is a variation of matrix-assisted laser desorption/ionization (MALDI). In MALDI, the sample is mixed with a matrix material and applied to a metal plate before irradiation by a laser, whereas in SELDI, proteins of interest in a sample become bound to a surface before MS analysis. The sample surface is a key component in the purification, desorption, and ionization of the sample. SELDI is typically used with time-of-flight (TOF) mass spectrometers and is used to detect proteins in tissue samples, blood, urine, or other clinical samples, however, SELDI technology can potentially be used in any application by simply modifying the sample surface.

MALDI imaging mass spectrometry (MALDI-IMS) is the use of matrix-assisted laser desorption ionization as a mass spectrometry imaging technique in which the sample, often a thin tissue section, is moved in two dimensions while the mass spectrum is recorded. Advantages, like measuring the distribution of a large amount of analytes at one time without destroying the sample, make it a useful method in tissue-based study.

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important method for the accurate mass determination and characterization of proteins, and a variety of methods and instrumentations have been developed for its many uses. Its applications include the identification of proteins and their post-translational modifications, the elucidation of protein complexes, their subunits and functional interactions, as well as the global measurement of proteins in proteomics. It can also be used to localize proteins to the various organelles, and determine the interactions between different proteins as well as with membrane lipids.
Protein footprinting is a term used to refer to a method of biochemical analysis that investigates protein structure, assembly, and interactions within a larger macromolecular assembly. It was originally coined in reference to the use of limited proteolysis to investigate contact sites within a monoclonal antibody - protein antigen complex and a year later to examine the protection from hydroxyl radical cleavage conferred by a protein bound to DNA within a DNA-protein complex. In DNA footprinting the protein is envisioned to make an imprint at a particular point of interaction. This latter method was adapted through the direct treatment of proteins and their complexes with hydroxyl radicals.
Kevin Downard is an Australian academic scientist whose research specialises in the improving responses to infectious disease through the application and development of mass spectrometry and other molecular approaches in the life and medical sciences. Downard has 30 years of experience in the field and written over 100 scientific publications and two books including a textbook for the Royal Society of Chemistry and the first book to be published on the role of mass spectrometry in the study of protein interactions.
Mass spectrometric immunoassay (MSIA) is a rapid method is used to detect and/ or quantify antigens and or antibody analytes. This method uses an analyte affinity isolation to extract targeted molecules and internal standards from biological fluid in preparation for matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS). This method allows for "top down" and "bottom up" analysis. This This sensitive method allows for a new and improved process for detecting multiple antigens and antibodies in a single assay. This assay is also capable of distinguishing mass shifted forms of the same molecule via a panantibody, as well as distinguish point mutations in proteins. Each specific form is detected uniquely based on their characteristic molecular mass. MSIA has dual specificity because of the antibody-antigen reaction coupled with the power of a mass spectrometer.
Antigen-antibody interaction, or antigen-antibody reaction, is a specific chemical interaction between antibodies produced by B cells of the white blood cells and antigens during immune reaction. It is the fundamental reaction in the body by which the body is protected from complex foreign molecules, such as pathogens and their chemical toxins. In the blood, the antigens are specifically and with high affinity bound by antibodies to form an antigen-antibody complex. The immune complex is then transported to cellular systems where it can be destroyed or deactivated.
Stable isotope standards and capture by anti-peptide antibodies (SISCAPA) is a mass spectrometry method for measuring the amount of a protein in a biological sample.








