Related Research Articles

A bacteriophage, also known informally as a phage, is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.

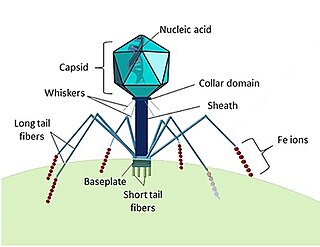
A capsid is the protein shell of a virus, enclosing its genetic material. It consists of several oligomeric (repeating) structural subunits made of protein called protomers. The observable 3-dimensional morphological subunits, which may or may not correspond to individual proteins, are called capsomeres. The proteins making up the capsid are called capsid proteins or viral coat proteins (VCP). The capsid and inner genome is called the nucleocapsid.
A cosmid is a type of hybrid plasmid that contains a Lambda phage cos sequence. They are often used as a cloning vector in genetic engineering. Cosmids can be used to build genomic libraries. They were first described by Collins and Hohn in 1978. Cosmids can contain 37 to 52 kb of DNA, limits based on the normal bacteriophage packaging size. They can replicate as plasmids if they have a suitable origin of replication (ori): for example SV40 ori in mammalian cells, ColE1 ori for double-stranded DNA replication, or f1 ori for single-stranded DNA replication in prokaryotes. They frequently also contain a gene for selection such as antibiotic resistance, so that the transformed cells can be identified by plating on a medium containing the antibiotic. Those cells which did not take up the cosmid would be unable to grow.

Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae from the family Myoviridae. T4 is capable of undergoing only a lytic life cycle and not the lysogenic life cycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains, Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.

Filamentous bacteriophages are a family of viruses (Inoviridae) that infect bacteria, or bacteriophages. They are named for their filamentous shape, a worm-like chain, about 6 nm in diameter and about 1000-2000 nm long. This distinctive shape reflects their method of replication: the coat of the virion comprises five types of viral protein, which are located in the inner membrane of the host bacterium during phage assembly, and these proteins are added to the nascent virion's DNA as it is extruded through the membrane. The simplicity of filamentous phages makes them an appealing model organism for research in molecular biology, and they have also shown promise as tools in nanotechnology and immunology.

M13 is one of the Ff phages, a member of the family filamentous bacteriophage (inovirus). Ff phages are composed of circular single-stranded DNA (ssDNA), which in the case of the m13 phage is 6407 nucleotides long and is encapsulated in approximately 2700 copies of the major coat protein p8, and capped with about 5 copies each of four different minor coat proteins. The minor coat protein p3 attaches to the receptor at the tip of the F pilus of the host Escherichia coli. The life cycle is relatively short, with the early phage progeny exiting the cell ten minutes after infection. Ff phages are chronic phage, releasing their progeny without killing the host cells. The infection causes turbid plaques in E. coli lawns, of intermediate opacity in comparison to regular lysis plaques. However, a decrease in the rate of cell growth is seen in the infected cells. M13 plasmids are used for many recombinant DNA processes, and the virus has also been used for phage display, directed evolution, nanostructures and nanotechnology applications.

The phi X 174 bacteriophage is a single-stranded DNA (ssDNA) virus that infects Escherichia coli, and the first DNA-based genome to be sequenced. This work was completed by Fred Sanger and his team in 1977. In 1962, Walter Fiers and Robert Sinsheimer had already demonstrated the physical, covalently closed circularity of ΦX174 DNA. Nobel prize winner Arthur Kornberg used ΦX174 as a model to first prove that DNA synthesized in a test tube by purified enzymes could produce all the features of a natural virus, ushering in the age of synthetic biology. In 1972–1974, Jerard Hurwitz, Sue Wickner, and Reed Wickner with collaborators identified the genes required to produce the enzymes to catalyze conversion of the single stranded form of the virus to the double stranded replicative form. In 2003, it was reported by Craig Venter's group that the genome of ΦX174 was the first to be completely assembled in vitro from synthesized oligonucleotides. The ΦX174 virus particle has also been successfully assembled in vitro. In 2012, it was shown how its highly overlapping genome can be fully decompressed and still remain functional.
Bacteriophages (phages), potentially the most numerous "organisms" on Earth, are the viruses of bacteria. Phage ecology is the study of the interaction of bacteriophages with their environments.

Cyanophages are viruses that infect cyanobacteria, also known as Cyanophyta or blue-green algae. Cyanobacteria are a phylum of bacteria that obtain their energy through the process of photosynthesis. Although cyanobacteria metabolize photoautotrophically like eukaryotic plants, they have prokaryotic cell structure. Cyanophages can be found in both freshwater and marine environments. Marine and freshwater cyanophages have icosahedral heads, which contain double-stranded DNA, attached to a tail by connector proteins. The size of the head and tail vary among species of cyanophages. Cyanophages infect a wide range of cyanobacteria and are key regulators of the cyanobacterial populations in aquatic environments, and may aid in the prevention of cyanobacterial blooms in freshwater and marine ecosystems. These blooms can pose a danger to humans and other animals, particularly in eutrophic freshwater lakes. Infection by these viruses is highly prevalent in cells belonging to Synechococcus spp. in marine environments, where up to 5% of cells belonging to marine cyanobacterial cells have been reported to contain mature phage particles.

The mobilome is the entire set of mobile genetic elements in a genome. Mobilomes are found in eukaryotes, prokaryotes, and viruses. The compositions of mobilomes differ among lineages of life, with transposable elements being the major mobile elements in eukaryotes, and plasmids and prophages being the major types in prokaryotes. Virophages contribute to the viral mobilome.

Bacteriophage Qbeta, commonly referred to as Qbeta or Qβ, is a positive-strand RNA virus which infects bacteria that have F-pili, most commonly Escherichia coli. Its linear genome is packaged into an icosahedral capsid with a diameter of 28 nm. Bacteriophage Qβ enters its host cell after binding to the side of the F-pilus.
Host–parasite coevolution is a special case of coevolution, where a host and a parasite continually adapt to each other. This can create an evolutionary arms race between them. A more benign possibility is of an evolutionary trade-off between transmission and virulence in the parasite, as if it kills its host too quickly, the parasite will not be able to reproduce either. Another theory, the Red Queen hypothesis, proposes that since both host and parasite have to keep on evolving to keep up with each other, and since sexual reproduction continually creates new combinations of genes, parasitism favours sexual reproduction in the host.
Rauchvirus is a genus of viruses in the order Caudovirales, in the family Podoviridae. Bacteria serve as natural hosts. The genus contains only one species: Bordetella virus BPP1.
A prohead or procapsid is an immature viral capsid structure formed in the early stages of self-assembly of some bacteriophages, including the Caudovirales or tailed bacteriophages. Production and assembly of stable proheads is an essential precursor to bacteriophage genome packaging; this packaging activity can be replicated in vitro. The prohead structure may take a different shape from the head of a mature virion, as seen with the prohead of Bacillus subtilis phage φ29.
Bacteriophage Mu, also known as mu phage or mu bacteriophage, is a muvirus of the family Myoviridae which has been shown to cause genetic transposition. It is of particular importance as its discovery in Escherichia coli by Larry Taylor was among the first observations of insertion elements in a genome. This discovery opened up the world to an investigation of transposable elements and their effects on a wide variety of organisms. While Mu was specifically involved in several distinct areas of research, the wider implications of transposition and insertion transformed the entire field of genetics.

Virome refers to the assemblage of viruses that is often investigated and described by metagenomic sequencing of viral nucleic acids that are found associated with a particular ecosystem, organism or holobiont. The word is frequently used to describe environmental viral shotgun metagenomes. Viruses, including bacteriophages, are found in all environments, and studies of the virome have provided insights into nutrient cycling, development of immunity, and a major source of genes through lysogenic conversion. Also, the human virome has been characterized in nine organs of 31 Finnish individuals using qPCR and NGS methodologies.

The "Kill the Winner" hypothesis (KtW) is an ecological model of population growth involving prokaryotes, viruses and protozoans that links trophic interactions to biogeochemistry. The model is related to the Lotka–Volterra equations. It assumes that prokaryotes adopt one of two strategies when competing for limited resources: priority is either given to population growth ("winners") or survival ("defenders"). As "winners" become more abundant and active in their environment, their contact with host-specific viruses increases, making them more susceptible to viral infection and lysis. Thus, viruses moderate the population size of "winners" and allow multiple species to coexist. Current understanding of KtW primarily stems from studies of lytic viruses and their host populations.
Auxiliary metabolic genes (AMGs) are found in many bacteriophages but originated in bacterial cells. AMGs modulate host cell metabolism during infection so that the phage can replicate more efficiently. For instance, bacteriophages that infect the abundant marine cyanobacteria Synechococcus and Prochlorococcus (cyanophages) carry AMGs that have been acquired from their immediate host as well as more distantly-related bacteria. Cyanophage AMGs support a variety of functions including photosynthesis, carbon metabolism, nucleic acid synthesis and metabolism.
Marine viruses are defined by their habitat as viruses that are found in marine environments, that is, in the saltwater of seas or oceans or the brackish water of coastal estuaries. Viruses are small infectious agents that can only replicate inside the living cells of a host organism, because they need the replication machinery of the host to do so. They can infect all types of life forms, from animals and plants to microorganisms, including bacteria and archaea.
Venigalla Basaveswara Rao is an Indian-American biochemist who is a professor of biology at the Catholic University of America. He serves as Director of the Bacteriophage Medical Research Center. In 2021, he was elected a Fellow of the American Society for Microbiology and the National Academy of Inventors.
References
- ↑ Miller, Leslie (5 July 2023). "Joshua Weitz Joins Biology as Professor and Clark Leadership Chair in Data Analytics". News. University of Maryland. Retrieved 12 September 2023.
- ↑ "Joshua Weitz". gatech.edu. Retrieved December 22, 2017.
- ↑ "Director Joshua Weitz welcomes the inaugural class to the QBioS Ph.D. at Georgia Tech". August 31, 2016. Retrieved September 10, 2018.
- ↑ "Joshua Weitz Elected AAAS Fellow". gatech.edu. November 21, 2017. Retrieved September 10, 2018.
- 1 2 "Joshua Weitz". gatech.edu. Retrieved December 22, 2017.
- ↑ Weitz, Joshua (January 5, 2016). Quantitative Viral Ecology: Dynamics of Viruses and Their Microbial Hosts. Princeton University Press. ISBN 978-0691161549.
- ↑ "Book Awards Winners 2016". Royal Society of Biology. Retrieved 10 September 2018.
- ↑ P.S. Dodds; D.H. Rothman; J.S. Weitz (7 March 2001). "Re-examination of the "3/4-law" of Metabolism". Journal of Theoretical Biology. 209 (1): 9–27. arXiv: physics/0007096 . Bibcode:2001JThBi.209....9D. doi:10.1006/jtbi.2000.2238. PMID 11237567. S2CID 9168199.
- ↑ J. S. Weitz; H. Hartman; S.A. Levin (July 5, 2005). "Coevolutionary arms races between bacteria and bacteriophage". Proceedings of the National Academy of Sciences. 102 (27): 9535–9540. Bibcode:2005PNAS..102.9535W. doi: 10.1073/pnas.0504062102 . PMC 1172273 . PMID 15976021.
- ↑ Cesar O. Flores; Justin R. Meyer; Sergi Valverde; Lauren Farr; Joshua S. Weitz (July 12, 2011). "Statistical structure of host–phage interaction". Proceedings of the National Academy of Sciences. 108 (28): E288–E297. doi: 10.1073/pnas.1101595108 . PMC 3136311 . PMID 21709225.
- ↑ Joshua S. Weitz; Steven W. Wilhelm (September 5, 2012). "Ocean viruses and their effects on microbial communities and biogeochemical cycles". F1000 Biol. Rep. 4 (17): 17. doi: 10.3410/B4-17 . PMC 3434959 . PMID 22991582.
- ↑ Li Deng; et al. (11 September 2014). "Viral tagging reveals discrete populations in Synechococcus viral genome sequence space". Nature. 513 (7517): 242–245. Bibcode:2014Natur.513..242D. doi:10.1038/nature13459. PMID 25043051. S2CID 4463116.
- ↑ Joshua S. Weitz; Ceyhun Eksin; Keith Paarporn; Sam P. Brown; William C. Ratcliff (November 22, 2016). "An oscillating tragedy of the commons in replicator dynamics with game-environment feedback". Proceedings of the National Academy of Sciences. 113 (47): E7518–E7525. Bibcode:2016PNAS..113E7518W. doi: 10.1073/pnas.1604096113 . PMC 5127343 . PMID 27830651.
- ↑ Between Two Stones: Poems. Sheep Meadow. November 1, 2002. ISBN 1931357021.
- ↑ Weitz, Joshua (March 5, 2017). "Should Scientists Compromise? First, Define Your Terms". The Chronicle for Higher Education. No. March 10, 2017. Retrieved 10 September 2018.
- ↑ Hagen, Lisa (April 24, 2017). "Thousands, Armed With Puns, March For Science In Atlanta". WABE. Retrieved 10 September 2018.