
In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, catalyze reactions, or to bind other RNA or protein macromolecules.

Lactococcus lactis is a gram-positive bacterium used extensively in the production of buttermilk and cheese, but has also become famous as the first genetically modified organism to be used alive for the treatment of human disease. L. lactis cells are cocci that group in pairs and short chains, and, depending on growth conditions, appear ovoid with a typical length of 0.5 - 1.5 µm. L. lactis does not produce spores (nonsporulating) and are not motile (nonmotile). They have a homofermentative metabolism, meaning they produce lactic acid from sugars. They've also been reported to produce exclusive L-(+)-lactic acid. However, reported D-(−)-lactic acid can be produced when cultured at low pH. The capability to produce lactic acid is one of the reasons why L. lactis is one of the most important microorganisms in the dairy industry. Based on its history in food fermentation, L. lactis has generally recognized as safe (GRAS) status, with few case reports of it being an opportunistic pathogen.

The Nirenberg and Leder experiment was a scientific experiment performed in 1964 by Marshall W. Nirenberg and Philip Leder. The experiment elucidated the triplet nature of the genetic code and allowed the remaining ambiguous codons in the genetic code to be deciphered.

The asd RNA motif is a conserved RNA structure found in certain lactic acid bacteria. The asd motif was detected by bioinformatics and an individual asd RNA in Streptococcus pyogenes was detected by microarray and northern hybridization experiments as a 170-nucleotide molecule called "SR914400". The transcription start site determined for SR914400 corresponds to the 5′-end of the molecule shown in the consensus diagram.

The Bacteroid-trp RNA motif is a conserved RNA element detected by bioinformatics. It is found in the phylum Bacteroidota in the apparent 5' untranslated regions of genes that encode enzymes used in the synthesis of the amino acid tryptophan. A short open reading frame is found within the motif that encodes at least two tryptophan codons. Similar motifs have been identified regulating tryptophan genes in Pseudomonadota, but not in Bacteroidota. However, the Bacteroid-trp RNA motif likely operates via the same mechanism of attenuation.

The Downstream-peptide motif refers to a conserved RNA structure identified by bioinformatics in the cyanobacterial genera Synechococcus and Prochlorococcus and one phage that infects such bacteria. It was also detected in marine samples of DNA from uncultivated bacteria, which are presumably other species of cyanobacteria.

The glutamine riboswitch is a conserved RNA structure that was predicted by bioinformatics. It is present in a variety of lineages of cyanobacteria, as well as some phages that infect cyanobacteria. It is also found in DNA extracted from uncultivated bacteria living in the ocean that are presumably species of cyanobacteria.
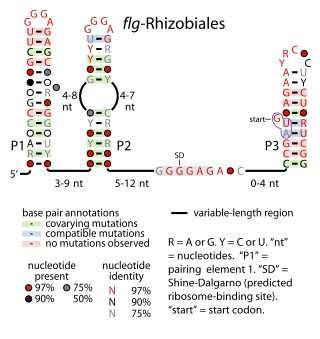
The flg-Rhizobiales RNA motif is an RNA structure that is conserved in certain bacteria. All known flg-Rhizobiales RNAs are located in the presumptive 5' untranslated regions of operons that contain genes whose functions relate to the creation of flagellar basal bodies. The flg-Rhizobiales RNAs are restricted to the Hyphomicrobiales, an order of alphaproteobacteria, although only some Rhizobiales bacterial are predicted to use flg-Rhizobiales RNAs. The exact function of these RNAs is unknown, although it is hypothesized that they have a cis-regulatory function in controlling expression of the downstream operons.

The gyrA RNA motif is a conserved RNA structure identified by bioinformatics. The RNAs are present in multiple species of bacteria within the order Pseudomonadales. This order contains the genus Pseudomonas, which includes the opportunistic human pathogen Pseudomonas aeruginosa and Pseudomonas syringae, a plant pathogen.

The L17 downstream element RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. All known L17 downstream elements were detected immediately downstream of genes encoding the L17 subunit of the ribosome, and therefore might be in the 3' untranslated regions of these genes. The element is found in a variety of lactic acid bacteria and in the genus Listeria.

The lactis-plasmid RNA motif is a conserved RNA structure identified by bioinformatics. The RNAs are restricted to lactic acid bacteria, and are especially common in Lactococcus lactis. They typically lie near to repB genes, and are almost found in plasmids. This data suggested that lactis-plasmid RNAs participate in the control of plasmid abundance. However, many of the plasmids that carry lactis-plasmid RNAs also carry ctRNA-pND324 RNAs, which are involved in plasmid copy count regulation. Therefore lactis-plasmid RNAs might have a different function.
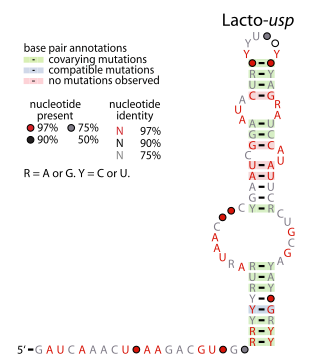
The Lacto-usp RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. Lacto-usp RNAs are found exclusively in lactic acid bacteria, and exclusively in the possible 5′ untranslated regions of operons that contain a hypothetical gene and a usp gene. The usp gene encodes the universal stress protein. It was proposed that the Lacto-usp might correspond to the 6S RNA of the relevant species, because four of five of these species lack a predicted 6S RNA, and 6S RNAs commonly occur in 5′ UTRs of usp genes. However, given that the Lacto-usp RNA motif is much shorter than the standard 6S RNA structure, the function of Lacto-usp RNAs remains unclear.

The livK RNA motif describes a conserved RNA structure that was discovered using bioinformatics. The livK motif is detected only in the species Pseudomonas syringae. It is found in the potential 5' untranslated regions of livK genes and downstream livM and livH genes, as well as the 5' UTRs of amidase genes. The liv genes are predicted to be transporters of branched-chain amino acids, i.e., leucine, isoleucine or valine. The specific reaction catalyzed the amidase genes is not predicted.
The Lnt RNA motif refers to a conserved RNA structure found in certain bacteria. Specifically, Lnt RNAs are known only in species within the phylum Chlorobiota, and are located in the possible 5' untranslated regions of genes that are annotated as encoding apolipoprotein N-acyltransferase enzymes. There is some doubt as to whether the indicated motif is transcribed as RNA, or whether its reverse complement is transcribed. If the reverse complement is transcribed it would potentially in 5' UTRs of genes encoding bacteriochlorophyll A, and would be close to the start codon of those genes.

The Ssbp, Topoisomerase, Antirestriction, XerDC Integrase RNA motif is a conserved RNA-like structure identified using bioinformatics. STAXI RNAs are located near to genes encoding proteins that interact with DNA or are associated with such proteins. This observation raised the possibility that instances of the STAXI motif function as single-stranded DNA molecules, perhaps during DNA replication or DNA repair. On the other hand, STAXI motifs often contain terminal loops conforming to the stable UNCG tetraloop, but the DNA version of this tetraloop (TNCG) is not especially stable. The STAXI motif consists of a simple pseudoknot structure that is repeated two or more times.
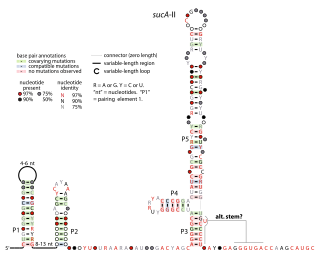
The sucA-II RNA motif is a conserved RNA structure identified by bioinformatics. It is consistently found in the presumed 5' untranslated regions of sucA genes, which encode Oxoglutarate dehydrogenase enzymes that participate in the citric acid cycle. Given this arrangement, sucA-II RNAs might regulate the downstream sucA gene. This genetic arrangement is similar to the previously reported sucA RNA motif. However, sucA-II RNAs are found only in bacteria classified within the genus Pseudomonas, whereas the previously reported motif is found only in betaproteobacteria.
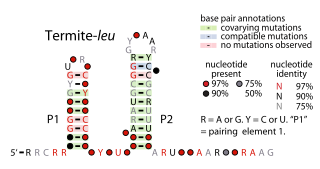
The Termite-leu RNA motif is a conserved RNA structure discovered by bioinformatics. It is found only in DNA sequences extracted from uncultivated bacteria living in termite hindguts, and has not yet been detected in any known cultivated organism. In many cases, Termite-leu RNAs are found in the likely 5′ untranslated regions of multive genes related to the synthesis of the amino acid leucine. However, in several cases it is not found in this type of location. Therefore, it was considered ambiguous as to whether Termite-leu RNAs constitute cis-regulatory elements.
The chlorophycean mitochondrial code is a genetic code found in the mitochondria of Chlorophyceae.
The leuA-Halobacteria RNA motif is a conserved RNA structure that was discovered by bioinformatics. leuA-Halobacteria motifs are found in Halobacteriaceae, a lineage of archaea.

The uup RNA motif is a conserved RNA structure that was discovered by bioinformatics. uup motif RNAs are found in Bacillota and Gammaproteobacteria.

















