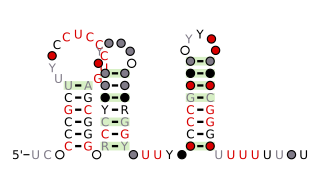
The anti-hemB RNA motif is a conserved RNA structure that was found in all known bacteria in the genus Burkholderia, and in a variety of other betaproteobacteria. The anti-hemB RNA motif consists primarily of two stem-loops, followed by a predicted rho-independent transcription termination stem-loop. As anti-hemB RNAs are generally not located in a 5' UTR, the RNAs are presumed to be non-coding RNAs. The terminator stem-loop implies that anti-hemB RNAs are transcribed as independent molecules.

The asd RNA motif is a conserved RNA structure found in certain lactic acid bacteria. The asd motif was detected by bioinformatics and an individual asd RNA in Streptococcus pyogenes was detected by microarray and northern hybridization experiments as a 170-nucleotide molecule called "SR914400". The transcription start site determined for SR914400 corresponds to the 5′-end of the molecule shown in the consensus diagram.

The Bacteroidales-1 RNA motif is a conserved RNA structure identified by bioinformatics. It has been identified only in bacteria within the order (biology) Bacteroidales. Its presumed length is marked by a promoter on one end that conforms to an alternate consensus sequence that is common in the phylum Bacteroidota, and its 3′ end is indicated by predicted transcription terminators. It is often located downstream of a gene that encodes the L20 ribosomal subunit, although it is unclear whether there is a functional reason underlying this apparent association.

The Bacteroides-1 RNA motif is a conserved RNA structure identified in bacteria within the genus Bacteroides. The RNAs are typically found downstream of of genes that participate in the synthesis of exopolysaccharides of unknown types. It is possible that Bacteroides-1 RNAs regulate the upstream genes, but since this mode of regulation is unusual in bacteria, it is more likely that the structure functions as a non-coding RNA.
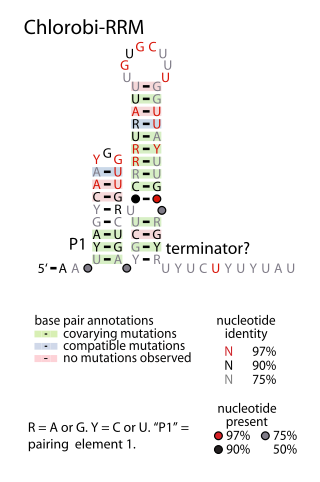
The Chlorobi-RRM RNA motif is a conserved RNA structure identified by bioinformatics. It is found within bacteria in the phylum Chlorobiota, and is exclusively detected in the presumed 5' untranslated regions of genes that encode putative RNA-binding proteins. Since many RNA-binding proteins regulate their own expression in a feedback mechanism by binding or acting up their 5' UTR, it was proposed that the Chlorobi-RRM is a component in an analogous feedback mechanism. Structurally, the motif consists of two stem-loops, the second of which might function as a rho-independent transcription terminator.

The Downstream-peptide motif refers to a conserved RNA structure identified by bioinformatics in the cyanobacterial genera Synechococcus and Prochlorococcus and one phage that infects such bacteria. It was also detected in marine samples of DNA from uncultivated bacteria, which are presumably other species of cyanobacteria.

The L17 downstream element RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. All known L17 downstream elements were detected immediately downstream of genes encoding the L17 subunit of the ribosome, and therefore might be in the 3' untranslated regions of these genes. The element is found in a variety of lactic acid bacteria and in the genus Listeria.
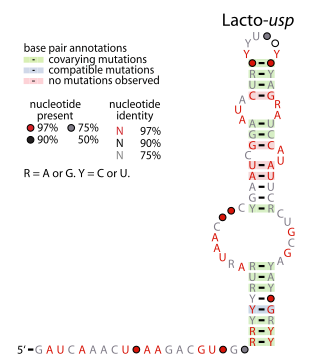
The Lacto-usp RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. Lacto-usp RNAs are found exclusively in lactic acid bacteria, and exclusively in the possible 5′ untranslated regions of operons that contain a hypothetical gene and a usp gene. The usp gene encodes the universal stress protein. It was proposed that the Lacto-usp might correspond to the 6S RNA of the relevant species, because four of five of these species lack a predicted 6S RNA, and 6S RNAs commonly occur in 5′ UTRs of usp genes. However, given that the Lacto-usp RNA motif is much shorter than the standard 6S RNA structure, the function of Lacto-usp RNAs remains unclear.

The livK RNA motif describes a conserved RNA structure that was discovered using bioinformatics. The livK motif is detected only in the species Pseudomonas syringae. It is found in the potential 5' untranslated regions of livK genes and downstream livM and livH genes, as well as the 5' UTRs of amidase genes. The liv genes are predicted to be transporters of branched-chain amino acids, i.e., leucine, isoleucine or valine. The specific reaction catalyzed the amidase genes is not predicted.
The nuoG RNA motif is a conserved RNA structure detected by bioinformatics. It is located in the presumed 5' untranslated regions of nuoG genes. This gene and the downstream genes probably comprise an operon that encodes various subunits of ubiquinone reductase enzyme.

PhotoRC RNA motifs refer to conserved RNA structures that are associated with genes acting in the photosynthetic reaction centre of photosynthetic bacteria. Two such RNA classes were identified and called the PhotoRC-I and PhotoRC-II motifs. PhotoRC-I RNAs were detected in the genomes of some cyanobacteria. Although no PhotoRC-II RNA has been detected in cyanobacteria, one is found in the genome of a purified phage that infects cyanobacteria. Both PhotoRC-I and PhotoRC-II RNAs are present in sequences derived from DNA that was extracted from uncultivated marine bacteria.

The psbNH RNA motif describes a class of RNA molecules that have a conserved secondary structure. psbNH RNAs are always found between psbH and psbH genes, both of which are involved in the cyanobacterial photosystem II are transcribed in opposite orientations. It is unknown whether the biological psbNH RNA is as depicted in the diagram, or whether its reverse complement is the transcribed molecule. In either case, the RNA would be in the 5' untranslated region of a gene, either psbN or psbH, and likely a cis-regulatory element.
The Pseudomon-groES RNA motif is a conserved RNA structure identified in certain bacteria using bioinformatics. It is found in most species within the family Pseudomonadaceae, and is consistently located in the 5' untranslated regions of operons that contain groES genes. RNA transcripts of the groES genes in Pseudomonas aeruginosa where shown experimentally to be initiated at one of two start sites, from promoters called "P1" and "P2". The Pseudomon-groES RNA is in the 5' UTR of transcripts initiated from the P1 site, but is truncated in P2 transcripts. groES genes are involved in the cellular response to heat shock, but it is not thought that the Pseudomonas-groES RNA motif is involved in heat shock regulation. However, it is thought that the motif might regulate groES genes in response to other stimuli.

The rmf RNA motif is a conserved RNA structure that was originally detected using bioinformatics. rmf RNAs are consistently foundwithin species classified into the genus Pseudomonas, and is located potentially in the 5′ untranslated regions of rmf genes. These genes encodes the ribosome modulation factor protein, which affects the translation of genes by modifying ribosome structure in response to stress such as starvation. This ribosome modulation is a part of the stringent response in bacteria. The likely biological role of rmf RNAs is ambiguous. Since the RNA could be in the 5′ UTRs of protein-coding genes, it was hypothesized that it functions as a cis-regulatory element. This hypothesis is bolstered by the observation that ribosome modulation factor binds ribosomal RNA, and many cis-regulatory RNAs called ribosomal protein leaders participate in a feedback regulation mechanism by binding to proteins that normally bind to ribosomal RNA. However, since rmf RNAs are not very close to the rmf genes, they might function as non-coding RNAs.

The rne-II RNA motif is a conserved RNA structure identified using bioinformatics. It is detected only in species classified within the family Pseudomonadaceae, a group of gammaproteobacteria. rne-II RNAs are consistently located in the presumed 5' untranslated regions of genes that encode Ribonuclease E. The RNase E 5' UTR element is a previously identified RNA structure that is also found in the 5' UTRs of RNase E genes. However, the latter motif is found only in enterobacteria, and the two motifs have apparently unrelated structure. In view of their differences, it was hypothesized that rne-II RNAs fulfill the same functional role as RNase E 5' UTR elements, which is to regulate the levels of RNase E proteins by acting as a substrate for RNase E. Thus, when concentrations of RNase E are high, they will degrade their own messenger RNA.

The SAM-Chlorobi RNA motif is a conserved RNA structure that was identified by bioinformatics. The RNAs are found only in bacteria classified as within the phylum Chlorobiota. These RNAs are always in the 5' untranslated regions of operons that contain metK and ahcY genes. metK genes encode methionine adenosyltransferase, which synthesizes S-adenosyl methionine (SAM), and ahcY genes encode S-adenosylhomocysteine hydrolase, which degrade the related metabolite S-Adenosyl-L-homocysteine (SAH). In fact all predicted metK and ahcY genes within Chlorobiota bacteria as of 2010 are preceded by predicted SAM-Chlorobi RNAs. Predicted promoter sequences are consistently found upstream of SAM-Chlorobi RNAs, and these promoter sequences imply that SAM-Chlorobi RNAs are indeed transcribed as RNAs. The promoter sequences are commonly associated with strong transcription in the phyla Chlorobiota and Bacteroidota, but are not used by most lineages of bacteria. The placement of SAM-Chlorobi RNAs suggests that they are involved in the regulation of the metK/ahcY operon through an unknown mechanism.

The sucC RNA motif is a conserved RNA structure discovered using bioinformatics. sucC RNAs are found in the genus Pseudomonas. They ae consistently found in possible 5' untranslated regions of sucC genes. These genes encode Succinyl coenzyme A synthetase, and are hypothesised to be regulated by the sucC RNAs. sucC genes participate in the citric acid cycle, and another gene involved in the citric acid cycle, sucA, is also predicted to be regulated by a conserved RNA structure.

The traJ-II RNA motif is a conserved RNA structure discovered in bacteria by using bioinformatics. traJ-II RNAs appear to be in the 5' untranslated regions of protein-coding genes called traJ, which functions in the process of bacterial conjugation. A previously identified motif known as TraJ 5' UTR is also found upstream of traJ genes functions as the target of FinP antisense RNAs, so it is possible that traJ-II RNAs play a similar role as targets of an antisense RNA. However, some sequence features within the traJ-II RNA motif suggest that the biological RNA might be transcribed from the reverse-complement strand. Thus is it unclear whether traJ-II function as cis-regulatory elements. traJ-II RNAs are found in a variety of Pseudomonadota.

The yjdF RNA motif is a conserved RNA structure identified using bioinformatics. Most yjdF RNAs are located in bacteria classified within the phylum Bacillota. A yjdF RNA is found in the presumed 5' untranslated region of the yjdF gene in Bacillus subtilis, and almost all yjdF RNAs are found in the 5' UTRs of homologs of this gene. The function of the yjdF gene is unknown, but the protein that it is predicted to encode is classified by the Pfam Database as DUF2992.

The uup RNA motif is a conserved RNA structure that was discovered by bioinformatics. uup motif RNAs are found in Bacillota and Gammaproteobacteria.


















