The Locus of Enterocyte Effacement (LEE) is a 35.6-Kb pathogenicity island which contains the genetic information for producing virulence factors, and mediating the pathogenicity of Escherichia coli bacteria. First characterized in 19981, It contains a set of 41 open reading frames arranged into five polycistronic operons which are regulated by a master regulator called the LEE encoded regulator (LER)2,3,4. The LER regulates transcription of proteins and functional elements encoded on LEE2-LEE4, and is an H-NS paralogue located upstream of the LEE2-LEE4 operons at orf1 of the LEE5,6. The LEE is regulated transcriptionally and post-transcriptionally, as it has regulators both within and outside the locus7. LEE1-LEE3 code for components of the type three secretion system, while LEE4 encodes a family of effector proteins deemed “esp” proteins (which stands for E. coli secreted proteins) as well as several other virulence factors. Notably LEE also codes for the production of Intimin, an adherence factor, as well as a receptor protein (Tir) that binds it. Intimin is an adhesion molecule that is necessary for the attachment of E. Coli to the intestinal wall. Tir is translocated into the host plasma membrane via the T3SS, where it can than bind intimin and mediate the formation of A/E lesions8. The LEE is an actively studied regulator of virulence due to the presence of type-three secretion systems in many different gram negative bacteria. T3SS can be found in Yersenia, Salmonella, Pseudomonas and Escherichia bacterial strains9
Other proteins of note include intimin, Tir, EspC, EspF, EspH, and Map protein. The LEE has a 38% G+C ratio.
1. [1] 2. [2] 3. [3] 4. [4] 5. [5] 6. [6] 7. [7] 8. [8] 9. [9] 10. [10]
Pathogenicity islands (PAIs), as termed in 1990, are a distinct class of genomic islands acquired by microorganisms through horizontal gene transfer. Pathogenicity islands are found in both animal and plant pathogens. Additionally, PAIs are found in both gram-positive and gram-negative bacteria. They are transferred through horizontal gene transfer events such as transfer by a plasmid, phage, or conjugative transposon. Therefore, PAIs contribute to microorganisms' ability to evolve.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.

Intimin is a virulence factor (adhesin) of EPEC and EHEC E. coli strains. It is an attaching and effacing (A/E) protein, which with other virulence factors is necessary and responsible for enteropathogenic and enterohaemorrhagic diarrhoea.
Tir is an essential component in the adherence of the enteropathogenic Escherichia coli (EPEC) and enterohemorraghic Escherichia coli (EHEC) to the cells lining the small intestine. To aid attachment, both EPEC and EHEC possess the ability to reorganise the host cell actin cytoskeleton via the secretion of virulence factors. These factors are secreted directly into the cells using a Type three secretion system. One of the virulence factors secreted is the Translocated Intimin Receptor (Tir). Tir is a receptor protein encoded by the espE gene which is located on the locus of enterocyte effacement (LEE) pathogenicity island in EPEC strains. It is secreted into the host cell membranes and acts as a receptor for intimin which is found on the bacterial surface. Once Tir binds intimin, the bacterium is attached to the enterocyte surface.
fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.

The OmrA-B RNA gene family is a pair of homologous OmpR-regulated small non-coding RNA that was discovered in E. coli during two large-scale screens. OmrA-B is highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well. RygB is adjacent to RygA a closely related RNA. These RNAs bind to the Hfq protein and regulate gene expression by antisense binding. They negatively regulate the expression of several genes encoding outer membrane proteins, including cirA, CsgD, fecA, fepA and ompT by binding in the vicinity of the Shine-Dalgarno sequence, suggesting the control of these targets is dependent on Hfq protein and RNase E. Taken together, these data suggest that OmrA-B participates in the regulation of outer membrane composition, responding to environmental conditions.

In molecular biology the ArcZ RNA is a small non-coding RNA (ncRNA). It is the functional product of a gene which is not translated into protein. ArcZ is an Hfq binding RNA that functions as an antisense regulator of a number of protein coding genes.

The Hfq protein encoded by the hfq gene was discovered in 1968 as an Escherichia coli host factor that was essential for replication of the bacteriophage Qβ. It is now clear that Hfq is an abundant bacterial RNA binding protein which has many important physiological roles that are usually mediated by interacting with Hfq binding sRNA.
In biology, phase variation is a method for dealing with rapidly varying environments without requiring random mutation. It involves the variation of protein expression, frequently in an on-off fashion, within different parts of a bacterial population. As such the phenotype can switch at frequencies that are much higher than classical mutation rates. Phase variation contributes to virulence by generating heterogeneity. Although it has been most commonly studied in the context of immune evasion, it is observed in many other areas as well and is employed by various types of bacteria, including Salmonella species.
Membrane ruffling (also known as cell ruffling) is the formation of a motile cell surface that contains a meshwork of newly polymerized actin filaments. It can also be regarded as one of the earliest structural changes observed in the cell. The GTP-binding protein Rac is the regulator of this membrane ruffling. Changes in the Polyphosphoinositide metabolism and changes in Ca2+ level of the cell may also play an important role. A number of actin-binding and organizing proteins localize to membrane ruffles and potentially target to transducing molecules.
Enteroinvasive Escherichia coli (EIEC) is a type of pathogenic bacteria whose infection causes a syndrome that is identical to shigellosis, with profuse diarrhea and high fever. EIEC are highly invasive, and they use adhesin proteins to bind to and enter intestinal cells. They produce no toxins, but severely damage the intestinal wall through mechanical cell destruction.
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
Shigatoxigenic Escherichia coli (STEC) and verotoxigenic E. coli (VTEC) are strains of the bacterium Escherichia coli that produce either Shiga toxin or Shiga-like toxin (verotoxin). Only a minority of the strains cause illness in humans. The ones that do are collectively known as enterohemorrhagic E. coli (EHEC) and are major causes of foodborne illness. When infecting humans, they often cause gastroenteritis, enterocolitis, and bloody diarrhea and sometimes cause a severe complication called hemolytic-uremic syndrome (HUS). The group and its subgroups are known by various names. They are distinguished from other strains of intestinal pathogenic E. coli including enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), and diffusely adherent E. coli (DAEC).

In molecular biology, the ferric uptake regulator family is a family of bacterial proteins involved in regulating metal ion uptake and in metal homeostasis. The family is named for its founding member, known as the ferric uptake regulator or ferric uptake regulatory protein (Fur). Fur proteins are responsible for controlling the intracellular concentration of iron in many bacteria. Iron is essential for most organisms, but its concentration must be carefully managed over a wide range of environmental conditions; high concentrations can be toxic due to the formation of reactive oxygen species.
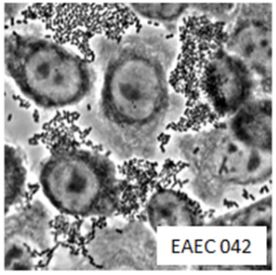
Enteroaggregative Escherichia coli are a pathotype of Escherichia coli is a cause of acute and chronic diarrhea in both the developed and developing world. They may also cause urinary tract infections. EAEC are defined by their "stacked-brick" pattern of adhesion to the human laryngeal epithelial cell line HEp-2. The pathogenesis of EAEC involves the aggregation of and adherence of the bacteria to the intestinal mucosa, where they elaborate enterotoxins and cytotoxins that damage host cells and induce inflammation that results in diarrhea.

Bacterial effectors are proteins secreted by pathogenic bacteria into the cells of their host, usually using a type 3 secretion system (TTSS/T3SS), a type 4 secretion system (TFSS/T4SS) or a Type VI secretion system (T6SS). Some bacteria inject only a few effectors into their host’s cells while others may inject dozens or even hundreds. Effector proteins may have many different activities, but usually help the pathogen to invade host tissue, suppress its immune system, or otherwise help the pathogen to survive. Effector proteins are usually critical for virulence. For instance, in the causative agent of plague, the loss of the T3SS is sufficient to render the bacteria completely avirulent, even when they are directly introduced into the bloodstream. Gram negative microbes are also suspected to deploy bacterial outer membrane vesicles to translocate effector proteins and virulence factors via a novel membrane vesicle trafficking secretory pathway, in order to modify their environment or attack/invade target cells, for example, at the host-pathogen interface.
The gene rpoN encodes the sigma factor sigma-54, a protein in Escherichia coli and other species of bacteria. RpoN antagonizes RpoS sigma factors.
Proteobiotics are natural metabolites which are produced by fermentation process of specific probiotic strains. These small oligopeptides were originally discovered in and isolated from culture media used to grow probiotic bacteria and may account for some of the health benefits of probiotics.
The locus of enterocyte effacement-encoded regulator (Ler) is a regulatory protein that controls bacterial pathogenicity of enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic Escherichia coli (EHEC). More specifically, Ler regulates the locus of enterocyte effacement (LEE) pathogenicity island genes, which are responsible for creating intestinal attachment and effacing lesions and subsequent diarrhea: LEE1, LEE2, and LEE3. LEE1, 2, and 3 carry the information necessary for a type III secretion system. The transcript encoding the Ler protein is the open reading frame 1 on the LEE1 operon.






