Bacterial conjugation is the transfer of genetic material between bacterial cells by direct cell-to-cell contact or by a bridge-like connection between two cells. This takes place through a pilus.

Escherichia coli O157:H7 is a serotype of the bacterial species Escherichia coli and is one of the Shiga toxin–producing types of E. coli. It is a cause of disease, typically foodborne illness, through consumption of contaminated and raw food, including raw milk and undercooked ground beef. Infection with this type of pathogenic bacteria may lead to hemorrhagic diarrhea, and to kidney failure; these have been reported to cause the deaths of children younger than five years of age, of elderly patients, and of patients whose immune systems are otherwise compromised.
A bacterial artificial chromosome (BAC) is a DNA construct, based on a functional fertility plasmid, used for transforming and cloning in bacteria, usually E. coli. F-plasmids play a crucial role because they contain partition genes that promote the even distribution of plasmids after bacterial cell division. The bacterial artificial chromosome's usual insert size is 150–350 kbp. A similar cloning vector called a PAC has also been produced from the DNA of P1 bacteriophage.

Shiga toxins are a family of related toxins with two major groups, Stx1 and Stx2, expressed by genes considered to be part of the genome of lambdoid prophages. The toxins are named after Kiyoshi Shiga, who first described the bacterial origin of dysentery caused by Shigella dysenteriae. Shiga-like toxin (SLT) is a historical term for similar or identical toxins produced by Escherichia coli. The most common sources for Shiga toxin are the bacteria S. dysenteriae and the some serotypes of Escherichia coli (STEC), which includes serotypes O157:H7, and O104:H4.
The nucleoid is an irregularly shaped region within the cell of a prokaryote that contains all or most of the genetic material, called genophore. In contrast to the nucleus of a eukaryotic cell, it is not surrounded by a nuclear membrane. The genome of prokaryotic organisms generally is a circular, double-stranded piece of DNA, of which multiple copies may exist at any time. The length of a genome widely varies, but generally is at least a few million base pairs. As in all cellular organisms, length of the DNA molecules of bacterial and archaeal chromosomes is very large compared to the dimensions of the cell, and the genomic DNA molecules must be compacted to fit.

Coliform bacteria are defined as rod-shaped Gram-negative non-spore forming and motile or non-motile bacteria which can ferment lactose with the production of acid and gas when incubated at 35–37°C. They are a commonly used indicator of sanitary quality of foods and water. Coliforms can be found in the aquatic environment, in soil and on vegetation; they are universally present in large numbers in the feces of warm-blooded animals. While coliforms themselves are not normally causes of serious illness, they are easy to culture, and their presence is used to indicate that other pathogenic organisms of fecal origin may be present. Such pathogens include disease-causing bacteria, viruses, or protozoa and many multicellular parasites. Coliform procedures are performed in aerobic or anaerobic conditions.
Enterotoxigenic Escherichia coli (ETEC) is a type of Escherichia coli and one of the leading bacterial causes of diarrhea in the developing world, as well as the most common cause of travelers' diarrhea. Insufficient data exist, but conservative estimates suggest that each year, about 157,000 deaths occur, mostly in children, from ETEC. A number of pathogenic isolates are termed ETEC, but the main hallmarks of this type of bacteria are expression of one or more enterotoxins and presence of fimbriae used for attachment to host intestinal cells. The bacteria was identified by the Bradley Sack lab in Kolkata in 1968.
Filamentation is the anomalous growth of certain bacteria, such as E. coli, in which cells continue to elongate but do not divide. The cells that result from elongation without division have multiple chromosomal copies. Bacterial filamentation is often observed as a result of bacteria responding to various stresses, including DNA damage or inhibition of replication. This may happen, for example, while responding to extensive DNA damage through the SOS response system. Nutritional changes may also cause bacterial filamentation. Some of the key genes involved in filamentation in E.coli include sulA and minCD. The following genes have been connected to virulence using the G. mellonella infection model: BCR1,FLO8, KEM1, SUV3 and TEC1. These genes are required for biofilm development from filamentation. Filamentation properties are argued to be necessary in virulence. The biofilm of bacteria is also connected to the organism’s virulence. Filamentation is a survival strategy that protects bacteria from stressors such as host effectors and protist predators. The strategy of filamentation is known to protect bacteria from antibiotic medicines taken by the host.
Pyrophosphatase is an enzyme that catalyzes the conversion of one molecule of pyrophosphate to two phosphate ions. This is a highly exergonic reaction, and therefore can be coupled to unfavorable biochemical transformations in order to drive these transformations to completion. The functionality of this enzyme plays a critical role in lipid metabolism, calcium absorption and bone formation, and DNA synthesis, as well as other biochemical transformations.
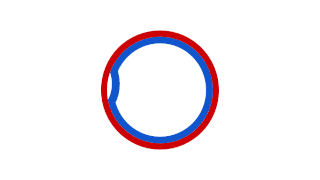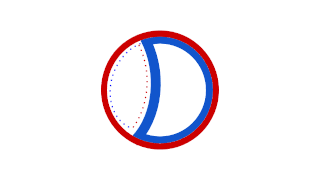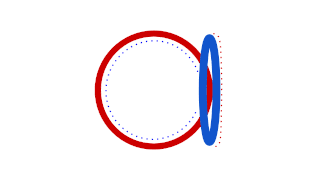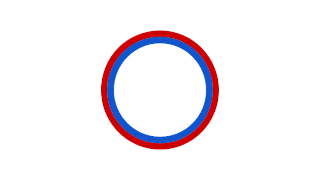
Prokaryotic DNA replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism E. coli, other bacteria show many similarities. Replication is bi-directional and originates at a single origin of replication (OriC). It consists of three steps: Initiation, elongation, and termination.
A P1-derived artificial chromosome is a DNA construct that was derived from the DNA of P1 bacteriophage. It can carry large amounts of other sequences for a variety of bioengineering purposes. It is one type of vector used to clone DNA fragments in Escherichia coli cells.

Tus, also known as terminus utilization substance, is a protein that binds to terminator sequences and acts as a counter-helicase when it comes in contact with an advancing helicase. The bound Tus protein effectively halts DNA polymerase movement. Tus helps end DNA replication in prokaryotes.

A circular prokaryote chromosome is a chromosome in bacteria and archaea, in the form of a molecule of circular DNA. Unlike the linear DNA of most eukaryotes, typical prokaryote chromosomes are circular.

In molecular biology the protein domain SeqA is one found in bacteria and archaea. The function of this protein domain is highly important in DNA replication. The protein negatively regulates the initiation of DNA replication at the origin of replication, in Escherichia coli, OriC. Additionally the protein plays a further role in sequestration. The importance of this protein is vital, without its help in DNA replication, cell division and other crucial processes could not occur. This protein domain is thought to be part of a much larger protein complex which includes other proteins such as SeqB.

In molecular biology, the ter site, also known as DNA replication terminus binding-site, refers to a protein domain which binds to the DNA replication terminus site. Ter-binding proteins are found in some bacterial species, and include the tus protein which is part of the common ter-tus binding domain. They are required for the termination of DNA replication and function by binding to DNA replication terminator sequences, thus preventing the passage of replication forks. The termination efficiency is affected by the affinity of a particular protein for the terminator sequence.
Élie Léo Wollman was a French microbial geneticist who first described plasmids, and served as vice director of research for the Pasteur Institute for twenty years. He was awarded the 1976 Grand Prix Charles-Leopold Mayer by the French Academy of Sciences and Chevalier of the French Legion of Honour.
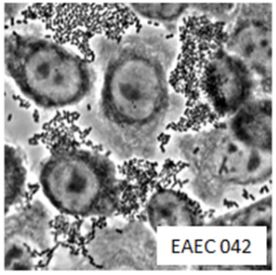
Enteroaggregative Escherichia coli are a pathotype of Escherichia coli associated with acute and chronic diarrhea in both the developed and developing world. EAEC are defined by their "stacked-brick" pattern of adhesion to the human laryngeal epithelial cell line HEp-2. The pathogenesis of EAEC involves the aggregation of and adherence of the bacteria to the intestinal mucosa, where they elaborate enterotoxins and cytotoxins that damage host cells and induce inflammation that results in diarrhea.
Maxicell is a system for identifying plasmid-encoded proteins that was developed by Sancar, AM Hack, and WD Rupp. The method uses a mutant strain of the bacterium Escherichia coli that is defective in repairing DNA damage. Irradiating the mutant cells with UV light leads to degradation of the bacterial chromosome. Plasmids in the cells mostly escape the UV-induced damage because of their small size. Proteins encoded by genes on the bacterial chromosome will no longer be synthesized, while proteins encoded by genes on the plasmid DNA molecules will continue to be made. The addition of radioactively-labeled amino acids following the UV treatment allows the plasmid-encoded proteins to be specifically visualized, because proteins synthesized prior to the UV treatment will not contain the radioactive label. The method was used for amplification of photolyase in Escherichia coli.









