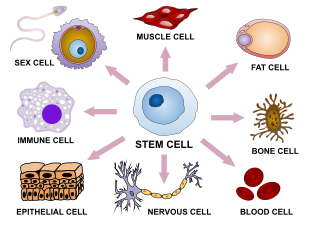
Cellular differentiation is the process in which a stem cell changes from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellular organism as it changes from a simple zygote to a complex system of tissues and cell types. Differentiation continues in adulthood as adult stem cells divide and create fully differentiated daughter cells during tissue repair and during normal cell turnover. Some differentiation occurs in response to antigen exposure. Differentiation dramatically changes a cell's size, shape, membrane potential, metabolic activity, and responsiveness to signals. These changes are largely due to highly controlled modifications in gene expression and are the study of epigenetics. With a few exceptions, cellular differentiation almost never involves a change in the DNA sequence itself. Metabolic composition, however, gets dramatically altered where stem cells are characterized by abundant metabolites with highly unsaturated structures whose levels decrease upon differentiation. Thus, different cells can have very different physical characteristics despite having the same genome.

Oct-4, also known as POU5F1, is a protein that in humans is encoded by the POU5F1 gene. Oct-4 is a homeodomain transcription factor of the POU family. It is critically involved in the self-renewal of undifferentiated embryonic stem cells. As such, it is frequently used as a marker for undifferentiated cells. Oct-4 expression must be closely regulated; too much or too little will cause differentiation of the cells.

GATA-binding factor 1 or GATA-1 is the founding member of the GATA family of transcription factors. This protein is widely expressed throughout vertebrate species. In humans and mice, it is encoded by the GATA1 and Gata1 genes, respectively. These genes are located on the X chromosome in both species.

Monoblasts are the committed progenitor cells that differentiated from a committed macrophage or dendritic cell precursor (MDP) in the process of hematopoiesis. They are the first developmental stage in the monocyte series leading to a macrophage. Their myeloid cell fate is induced by the concentration of cytokines they are surrounded by during development. These cytokines induce the activation of transcription factors which push completion of the monoblast's myeloid cell fate. Monoblasts are normally found in bone marrow and do not appear in the normal peripheral blood. They mature into monocytes which, in turn, develop into macrophages. They then are seen as macrophages in the normal peripheral blood and many different tissues of the body. Macrophages can produce a variety of effector molecules that initiate local, systemic inflammatory responses. These monoblast differentiated cells are equipped to fight off foreign invaders using pattern recognition receptors to detect antigen as part of the innate immune response.

Activator protein 1 (AP-1) is a transcription factor that regulates gene expression in response to a variety of stimuli, including cytokines, growth factors, stress, and bacterial and viral infections. AP-1 controls a number of cellular processes including differentiation, proliferation, and apoptosis. The structure of AP-1 is a heterodimer composed of proteins belonging to the c-Fos, c-Jun, ATF and JDP families.

Jun dimerization protein 2 (JUNDM2) is a protein that in humans is encoded by the JDP2 gene. The Jun dimerization protein is a member of the AP-1 family of transcription factors.

Induced pluripotent stem cells are a type of pluripotent stem cell that can be generated directly from a somatic cell. The iPSC technology was pioneered by Shinya Yamanaka and Kazutoshi Takahashi in Kyoto, Japan, who together showed in 2006 that the introduction of four specific genes, collectively known as Yamanaka factors, encoding transcription factors could convert somatic cells into pluripotent stem cells. Shinya Yamanaka was awarded the 2012 Nobel Prize along with Sir John Gurdon "for the discovery that mature cells can be reprogrammed to become pluripotent."

Tumor protein p63, typically referred to as p63, also known as transformation-related protein 63, is a protein that in humans is encoded by the TP63 gene.

GATA2 or GATA-binding factor 2 is a transcription factor, i.e. a nuclear protein which regulates the expression of genes. It regulates many genes that are critical for the embryonic development, self-renewal, maintenance, and functionality of blood-forming, lymphatic system-forming, and other tissue-forming stem cells. GATA2 is encoded by the GATA2 gene, a gene which often suffers germline and somatic mutations which lead to a wide range of familial and sporadic diseases, respectively. The gene and its product are targets for the treatment of these diseases.

Krüppel-like factor 4 is a member of the KLF family of zinc finger transcription factors, which belongs to the relatively large family of SP1-like transcription factors. KLF4 is involved in the regulation of proliferation, differentiation, apoptosis and somatic cell reprogramming. Evidence also suggests that KLF4 is a tumor suppressor in certain cancers, including colorectal cancer. It has three C2H2-zinc fingers at its carboxyl terminus that are closely related to another KLF, KLF2. It has two nuclear localization sequences that signals it to localize to the nucleus. In embryonic stem cells (ESCs), KLF4 has been demonstrated to be a good indicator of stem-like capacity. It is suggested that the same is true in mesenchymal stem cells (MSCs).

SRY -box 2, also known as SOX2, is a transcription factor that is essential for maintaining self-renewal, or pluripotency, of undifferentiated embryonic stem cells. Sox2 has a critical role in maintenance of embryonic and neural stem cells.

Transcription factor PU.1 is a protein that in humans is encoded by the SPI1 gene.

Transcription factor Spi-B is a protein that in humans is encoded by the SPIB gene.

Zinc finger and BTB domain-containing protein 32 is a protein that in humans is encoded by the 1960 bp ZBTB32 gene. The 52 kDa protein is a transcriptional repressor and the gene is expressed in T and B cells upon activation, but also significantly in testis cells. It is a member of the Poxviruses and Zinc-finger (POZ) and Krüppel (POK) family of proteins, and was identified in multiple screens involving either immune cell tumorigenesis or immune cell development.

Cell potency is a cell's ability to differentiate into other cell types. The more cell types a cell can differentiate into, the greater its potency. Potency is also described as the gene activation potential within a cell, which like a continuum, begins with totipotency to designate a cell with the most differentiation potential, pluripotency, multipotency, oligopotency, and finally unipotency.

Forkhead box protein A1 (FOXA1), also known as hepatocyte nuclear factor 3-alpha (HNF-3A), is a protein that in humans is encoded by the FOXA1 gene.
Pioneer factors are transcription factors that can directly bind condensed chromatin. They can have positive and negative effects on transcription and are important in recruiting other transcription factors and histone modification enzymes as well as controlling DNA methylation. They were first discovered in 2002 as factors capable of binding to target sites on nucleosomal DNA in compacted chromatin and endowing competency for gene activity during hepatogenesis. Pioneer factors are involved in initiating cell differentiation and activation of cell-specific genes. This property is observed in histone fold-domain containing transcription factors and other transcription factors that use zinc finger(s) for DNA binding.
H3K4me3 is an epigenetic modification to the DNA packaging protein Histone H3 that indicates tri-methylation at the 4th lysine residue of the histone H3 protein and is often involved in the regulation of gene expression. The name denotes the addition of three methyl groups (trimethylation) to the lysine 4 on the histone H3 protein.
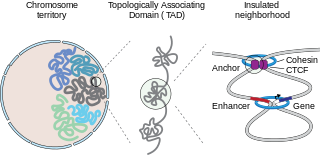
In mammalian biology, insulated neighborhoods are chromosomal loop structures formed by the physical interaction of two DNA loci bound by the transcription factor CTCF and co-occupied by cohesin. Insulated neighborhoods are thought to be structural and functional units of gene control because their integrity is important for normal gene regulation. Current evidence suggests that these structures form the mechanistic underpinnings of higher-order chromosome structures, including topologically associating domains (TADs). Insulated neighborhoods are functionally important in understanding gene regulation in normal cells and dysregulated gene expression in disease.

Thomas Graf is a biologist at the Centre for Genomic Regulation (CRG) in Barcelona, Spain. He is a pioneer in cell reprogramming, showing that blood cells can be transdifferentiated by transcription factors. He is also known for his early work on oncogenes carried by retroviruses and oncogene cooperation in leukemia formation.
















